J Liver Cancer.
2019 Mar;19(1):46-54. 10.17998/jlc.19.1.46.
Effect of PTEN Polymorphism on the Development of Hepatitis B Virus-associated Hepatocellular Carcinoma
- Affiliations
-
- 1Department of Gastroenterology, Ajou University School of Medicine, Suwon, Korea. jaeyoun620@gmail.com
- 2Clinical Trial Center, Ajou University Hospital, Suwon, Korea.
- 3Department of Biomedical Sciences, Ajou University Graduate School of Medicine, Suwon, Korea.
- KMID: 2448276
- DOI: http://doi.org/10.17998/jlc.19.1.46
Abstract
- BACKGROUND/AIMS
Phosphatase and tensin homolog (PTEN) is a known tumor suppressor gene that is downregulated in hepatocellular carcinoma (HCC). Here, we investigated the association between single nucleotide polymorphisms (SNPs) of PTEN and HCC development in patients with hepatitis B virus (HBV) infection.
METHODS
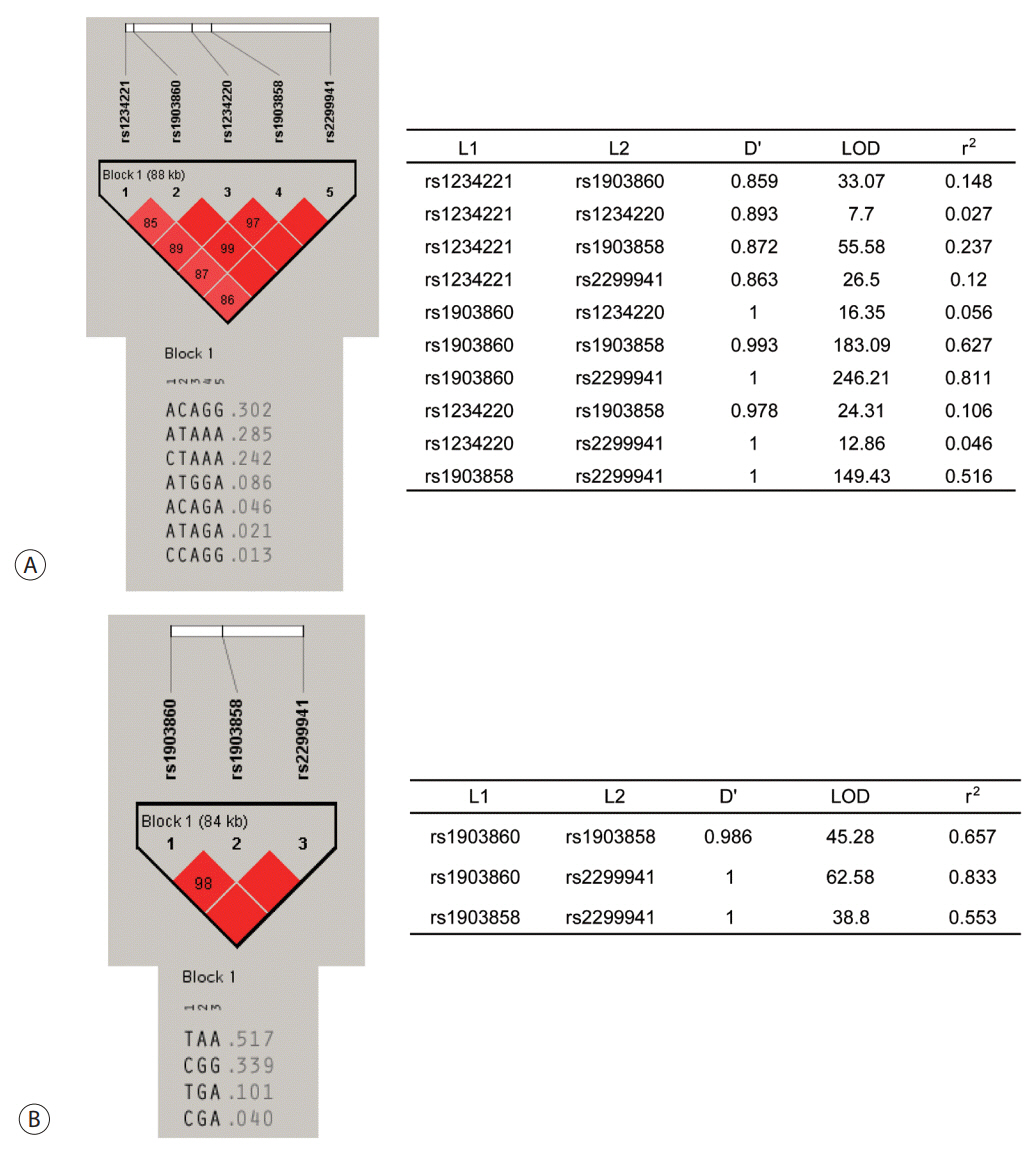
Six SNPs of PTEN at positions rs1234221, rs1903860, rs1234220, rs1903858, rs2299941, and rs17431184 were analyzed in a development population (417 chronic HBV carriers without HCC and 281 chronic HBV carriers with HCC). PTEN rs1903858, rs1903860, and rs2299941 SNPs were further assessed for the development of HCC in a validation population of 200 patients with HBV-related liver cirrhosis.
RESULTS
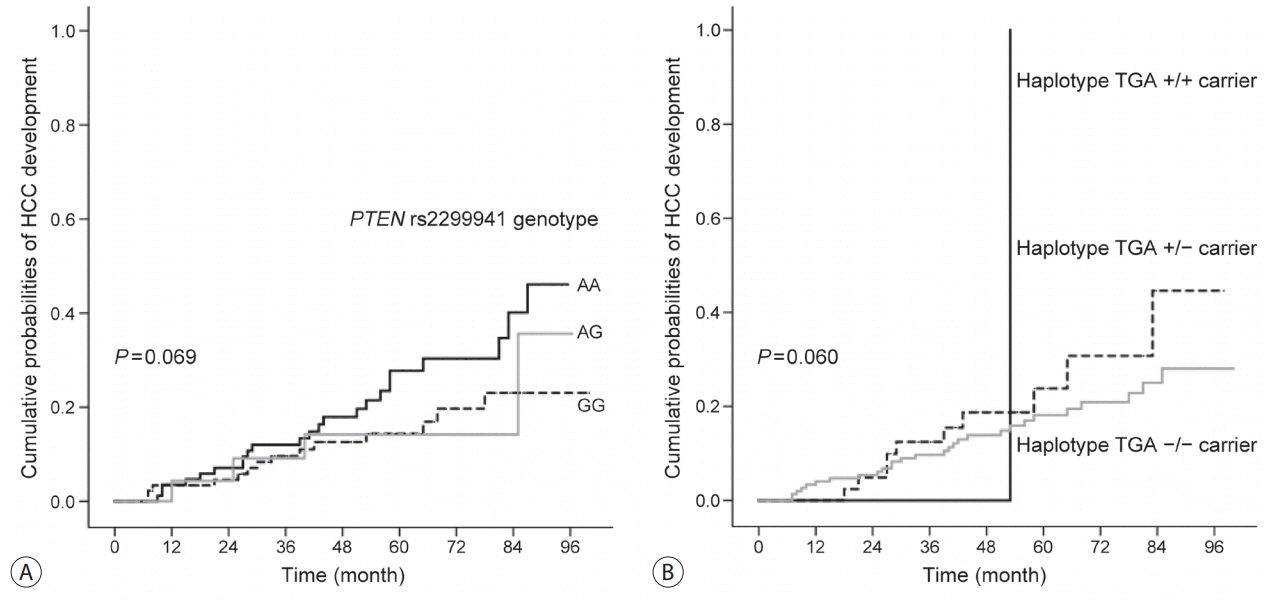
In the development population, PTEN rs1903860 C allele, rs1903858 G allele, and rs2299941 G allele were associated with a low risk of HCC. The haplotype A-T-A-A-A was associated with an increased risk of HCC (recessive model; odds ratio=2.277, 95% confidence interval [CI] =1.144-4.532, P=0.019). In the validation population, PTEN rs2299941 G allele was the only significant protective genetic polymorphism related to HCC development after adjustment for age and sex (hazard ratio=0.582, 95% CI =0.353-0.962, P=0.035).
CONCLUSIONS
These findings suggest that genetic polymorphisms in PTEN may affect HCC development in patients with chronic HBV infection.
MeSH Terms
Figure
Reference
-
1. Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014; 28:753–770.2. Lyu H, Lee D, Chung YH, Kim JA, Lee JH, Jin YJ, et al. Synergistic effects of A1896, T1653 and T1762/A1764 mutations in genotype c2 hepatitis B virus on development of hepatocellular carcinoma. J Viral Hepat. 2013; 20:219–224.3. Mathews P, Lee D, Chung YH, Kim JA, Lee JH, Jin YJ, et al. Effects of genomic changes in hepatitis B virus on postoperative recurrence and survival in patients with hepatocellular carcinoma. Ann Surg Oncol. 2013; 20:1216–1222.4. Jiang BH, Liu LZ. PI3K/PTEN signaling in tumorigenesis and angiogenesis. Biochim Biophys Acta. 2008; 1784:150–158.5. Li L, Ross AH. Why is PTEN an important tumor suppressor? J Cell Biochem. 2007; 102:1368–1374.6. Wang L, Wang WL, Zhang Y, Guo SP, Zhang J, Li QL. Epigenetic and genetic alterations of PTEN in hepatocellular carcinoma. Hepatol Res. 2007; 37:389–396.7. Khalid A, Hussain T, Manzoor S, Saalim M, Khaliq S. PTEN: a potential prognostic marker in virus-induced hepatocellular carcinoma. Tumour Biol. 2017; 39:1010428317705754.8. Kang-Park S, Im JH, Lee JH, Lee YI. PTEN modulates hepatitis B virus-X protein induced survival signaling in Chang liver cells. Virus Res. 2006; 122:53–60.9. Jin YJ, Lee D, Chung YH, Kim JA, Kim SE, Lee YS, et al. Tumor necrosis factor-alpha gene polymorphism associated with development of hepatitis B virus-associated hepatocellular carcinoma. J Clin Gastroenterol. 2015; 49:e76–e81.10. Kim SS, Cho HJ, Lee HY, Park JH, Noh CK, Shin SJ, et al. Genetic polymorphisms in the Wnt/beta-catenin pathway genes as predictors of tumor development and survival in patients with hepatitis B virus-associated hepatocellular carcinoma. Clin Biochem. 2016; 49:792–801.11. Liao X, Yu L, Liu X, Han C, Yu T, Qin W, et al. Genome-wide association pathway analysis to identify candidate single nucleotide polymorphisms and molecular pathways associated with TP53 expression status in HBV-related hepatocellular carcinoma. Cancer Manag Res. 2018; 10:953–967.12. Du Y, Zhang YW, Pu R, Han X, Hu JP, Zhang HW, et al. Phosphatase and tensin homologue genetic polymorphisms and their interactions with viral mutations on the risk of hepatocellular carcinoma. Chin Med J (Engl). 2015; 128:1005–1013.13. Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018; 68:723–750.14. Kim SS, Hwang JC, Lim SG, Ahn SJ, Cheong JY, Cho SW. Effect of virological response to entecavir on the development of hepatocellular carcinoma in hepatitis B viral cirrhotic patients: comparison between compensated and decompensated cirrhosis. Am J Gastroenterol. 2014; 109:1223–1233.15. Lee SK, Kim MH, Cheong JY, Cho SW, Yang SJ, Kwack K. Integrin alpha V polymorphisms and haplotypes in a Korean population are associated with susceptibility to chronic hepatitis and hepatocellular carcinoma. Liver Int. 2009; 29:187–195.16. Ding J, Gao Y, Liu R, Xu F, Liu H. Association of PTEN polymorphisms with susceptibility to hepatocellular carcinoma in a Han Chinese population. DNA Cell Biol. 2011; 30:229–234.17. Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006; 6:184–192.18. Rossi DJ, Weissman IL. PTEN, tumorigenesis, and stem cell selfrenewal. Cell. 2006; 125:229–231.19. Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004; 22:2954–2963.20. Wan XW, Jiang M, Cao HF, He YQ, Liu SQ, Qiu XH, et al. The alteration of PTEN tumor suppressor expression and its association with the histopathological features of human primary hepatocellular carcinoma. J Cancer Res Clin Oncol. 2003; 129:100–106.21. Dong-Dong L, Xi-Ran Z, Xiang-Rong C. Expression and significance of new tumor suppressor gene PTEN in primary liver cancer. J Cell Mol Med. 2003; 7:67–71.22. Wu SK, Wang BJ, Yang Y, Feng XH, Zhao XP, Yang DL. Expression of PTEN, PPM1A and P-Smad2 in hepatocellular carcinomas and adjacent liver tissues. World J Gastroenterol. 2007; 13:4554–4559.23. Xie CC, Lu L, Sun J, Zheng SL, Isaacs WB, Gronberg H, et al. Germline sequence variants of PTEN do not have an important role in hereditary and non-hereditary prostate cancer susceptibility. J Hum Genet. 2011; 56:496–502.24. Lacey JV Jr, Yang H, Gaudet MM, Dunning A, Lissowska J, Sherman ME, et al. Endometrial cancer and genetic variation in PTEN, PIK3CA, AKT1, MLH1, and MSH2 within a population-based casecontrol study. Gynecol Oncol. 2011; 120:167–173.25. Pezzolesi MG, Li Y, Zhou XP, Pilarski R, Shen L, Eng C. Mutationpositive and mutation-negative patients with Cowden and Bannayan-Riley-Ruvalcaba syndromes associated with distinct 10q haplotypes. Am J Hum Genet. 2006; 79:923–934.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A case of primary hepatocellular carcinoma following vertical transmission of hepatitis B virus in a child
- The use of transient elastography for predicting hepatocellular carcinoma in chronic hepatitis B patients: Editorial on “Risk assessment of hepatitis B virus-related hepatocellular carcinoma development using vibration-controlled transient elastography: Systematic review and meta-analysis”
- Serum Hepatitis B Virus DHA Level and Hepatocellulor Carcinoma
- Risk of hepatitis B virus-related hepatocellular carcinoma development is much higher in Koreans than in Taiwanese
- Hepatocellular Carcinoma Following Vertical Transmission of Hepatitis B Virus in a Child with X-linked Agammaglobulinemia