Korean J Neurotrauma.
2019 Apr;15(1):2-10. 10.13004/kjnt.2019.15.e2.
Molecular Characterization of Primary Human Astrocytes Using Digital Gene Expression Analysis
- Affiliations
-
- 1Department of Neurosurgery, Dongguk University Gyeongju Hospital, Dongguk University College of Medicine, Gyeongju, Korea. ktokhou@gmail.com
- KMID: 2444199
- DOI: http://doi.org/10.13004/kjnt.2019.15.e2
Abstract
OBJECTIVE
Astrocyte dysfunctions are related to several central nervous system (CNS) pathologies. Transcriptomic profiling of human mRNAs to investigate astrocyte functions may provide the basic molecular-biological data pertaining to the cellular activities of astrocytes.
METHODS
Human Primary astrocytes (HPAs) and human neural stem cell line (HB1.F3) were used for differential digital gene analysis. In this study, a massively parallel sequencing platform, next-generation sequencing (NGS), was used to obtain the digital gene expression (DGE) data from HPAs. A comparative analysis of the DGE from HPA and HB1.F3 cells was performed. Sequencing was performed using NGS platform, and subsequently, bioinformatic analyses were implemented to reveal the identity of the pathways, relatively up- or down-regulated in HPA cells.
RESULTS
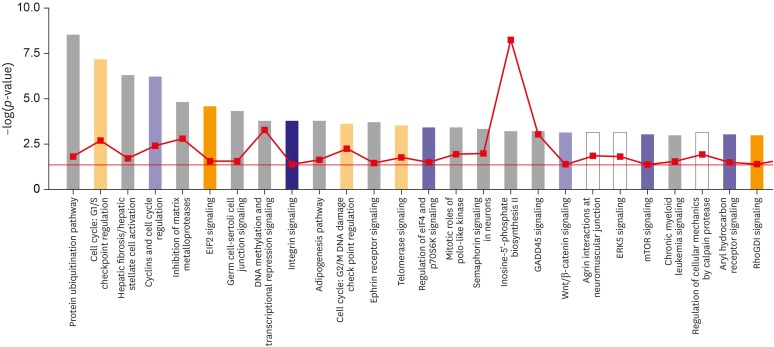
The top, novel canonical pathways up-regulated in HPA cells than in the HB1.F3 cells were "Cyclins and cell cycle regulation,""Integrin signaling,""Regulation of eIF4 and p70S6K signaling,""Wnt/β-catenin signaling,""mTOR signaling,""Aryl hydrocarbon receptor signaling,""Hippo signaling,""RhoA signaling,""Signaling by Rho family GTPases," and "Glioma signaling" pathways. The down-regulated pathways were "Cell cycle: G1/S checkpoint regulation,""eIF2 signaling,""Cell cycle: G2/M DNA damage checkpoint regulation,""Telomerase signaling,""RhoGDI signaling,""NRF2-mediated oxidative stress response,""ERK/MAPK signaling,""ATM signaling,""Pancreatic adenocarcinoma signaling,""VEGF signaling," and "Role of CHK proteins in cell cycle checkpoint control" pathways.
CONCLUSION
This study would be a good reference to understand astrocyte functions at the molecular level, and to develop a diagnostic test, based on the DGE pattern of astrocytes, as a powerful, new clinical tool in many CNS diseases.
Keyword
MeSH Terms
-
Adenocarcinoma
Astrocytes*
Cell Cycle
Cell Cycle Checkpoints
Central Nervous System
Central Nervous System Diseases
Computational Biology
Diagnostic Tests, Routine
DNA Damage
Gene Expression*
High-Throughput Nucleotide Sequencing
Humans*
Neural Stem Cells
Oxidative Stress
Pathology
Ribosomal Protein S6 Kinases, 70-kDa
RNA, Messenger
RNA, Messenger
Ribosomal Protein S6 Kinases, 70-kDa
Figure
Reference
-
1. Abbott NJ, Rönnbäck L, Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat Rev Neurosci. 7:41–53. 2006; PMID: 16371949.
Article2. Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 8:1765–1786. 2013; PMID: 23975260.
Article3. Banerjee S, Crouse NR, Emnett RJ, Gianino SM, Gutmann DH. Neurofibromatosis-1 regulates mTOR-mediated astrocyte growth and glioma formation in a TSC/Rheb-independent manner. Proc Natl Acad Sci U S A. 108:15996–16001. 2011; PMID: 21896734.
Article4. Benarroch EE. Neuron-astrocyte interactions: partnership for normal function and disease in the central nervous system. Mayo Clin Proc. 80:1326–1338. 2005; PMID: 16212146.
Article5. Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 18:630–634. 2000; PMID: 10835600.
Article6. Clemens MJ. Translational regulation in cell stress and apoptosis. Roles of the eIF4E binding proteins. J Cell Mol Med. 5:221–239. 2001; PMID: 12067482.
Article7. Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 149:1192–1205. 2012; PMID: 22682243.
Article8. Dazert E, Hall MN. mTOR signaling in disease. Curr Opin Cell Biol. 23:744–755. 2011; PMID: 21963299.
Article9. Dovas A, Couchman JR. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem J. 390:1–9. 2005; PMID: 16083425.
Article10. Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 106:489–498. 2001; PMID: 11525734.11. Fields RD, Stevens-Graham B. New insights into neuron-glia communication. Science. 298:556–562. 2002; PMID: 12386325.
Article12. Fukata M, Nakagawa M, Kaibuchi K. Roles of Rho-family GTPases in cell polarisation and directional migration. Curr Opin Cell Biol. 15:590–597. 2003; PMID: 14519394.
Article13. Hatton GI. Glial-neuronal interactions in the mammalian brain. Adv Physiol Educ. 26:225–237. 2002; PMID: 12443995.
Article14. Kiernan JA. Barr's the human nervous system: an anatomical viewpoint. ed 9. Philadelphia, PA: Lippincott Williams & Wilkins;2008.15. Kimelberg HK. Primary astrocyte cultures--a key to astrocyte function. Cell Mol Neurobiol. 3:1–16. 1983; PMID: 6136326.
Article16. Kimelberg HK, Bowman C, Biddlecome S, Bourke RS. Cation transport and membrane potential properties of primary astroglial cultures from neonatal rat brains. Brain Res. 177:533–550. 1979; PMID: 227541.
Article17. Kim SU. Human neural stem cells genetically modified for brain repair in neurological disorders. Neuropathology. . 24:159–171. 2004; PMID: 15484694.
Article18. Kim SU, de Vellis J. Stem cell-based cell therapy in neurological diseases: a review. J Neurosci Res. 87:2183–2200. 2009; PMID: 19301431.
Article19. Kim SU, Nagai A, Nakagawa E, Choi HB, Bang JH, Lee HJ, et al. Production and characterization of immortal human neural stem cell line with multipotent differentiation property. Methods Mol Biol. 438:103–121. 2008; PMID: 18369753.
Article20. Kim WK, Kim D, Cui J, Jang HH, Kim KS, Lee HJ, et al. Secretome analysis of human oligodendrocytes derived from neural stem cells. PLoS One. 9:e84292. 2014; PMID: 24392122.
Article21. Kuffler SW. Neuroglial cells: physiological properties and a potassium mediated effect of neuronal activity on the glial membrane potential. Proc R Soc Lond B Biol Sci. 168:1–21. 1967; PMID: 4382871.22. Nagai A, Nakagawa E, Hatori K, Choi HB, McLarnon JG, Lee MA, et al. Generation and characterization of immortalized human microglial cell lines: expression of cytokines and chemokines. Neurobiol Dis. 8:1057–1068. 2001; PMID: 11741401.
Article23. Pekny M, Nilsson M. Astrocyte activation and reactive gliosis. Glia. 50:427–434. 2005; PMID: 15846805.
Article24. Ridet JL, Malhotra SK, Privat A, Gage FH. Reactive astrocytes: cellular and molecular cues to biological function. Trends Neurosci. 20:570–577. 1997; PMID: 9416670.
Article25. Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 68:320–344. 2004; PMID: 15187187.
Article26. Singh I. Textbook of human neuroanatomy. ed 8. New Delhi: Jaypee Brothers Publishers;2006.27. Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 5:16–18. 2008; PMID: 18165802.
Article28. Vernadakis A. Neuron-glia interrelations. Int Rev Neurobiol. 30:149–224. 1988; PMID: 3061968.29. Vernadakis A. Glia-neuron intercommunications and synaptic plasticity. Prog Neurobiol. 49:185–214. 1996; PMID: 8878303.
Article30. Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 27:355–371. 2013; PMID: 23431053.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Characterization of Gene Expression Pattern in Human Astrocytes using DDRT - PCR Method
- Microarray Analysis of Oxygen-Glucose-Deprivation Induced Gene Expression in Cultured Astrocytes
- Regulation of TNF - alpha Gene Expression in Human Fetal Astrocytes
- Establishment of a Culture Method and Characterization for Human Fetal Astrocytes
- Arctigenin Increases Hemeoxygenase-1 Gene Expression by Modulating PI3K/AKT Signaling Pathway in Rat Primary Astrocytes