Ann Lab Med.
2012 Nov;32(6):433-437.
A Case of Helicobacter cinaedi Bacteremia in an Asplenic Patient
- Affiliations
-
- 1Department of Laboratory Medicine, University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea. mnkim@amc.seoul.kr
- 2Department of Internal Medicine, University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea.
Abstract
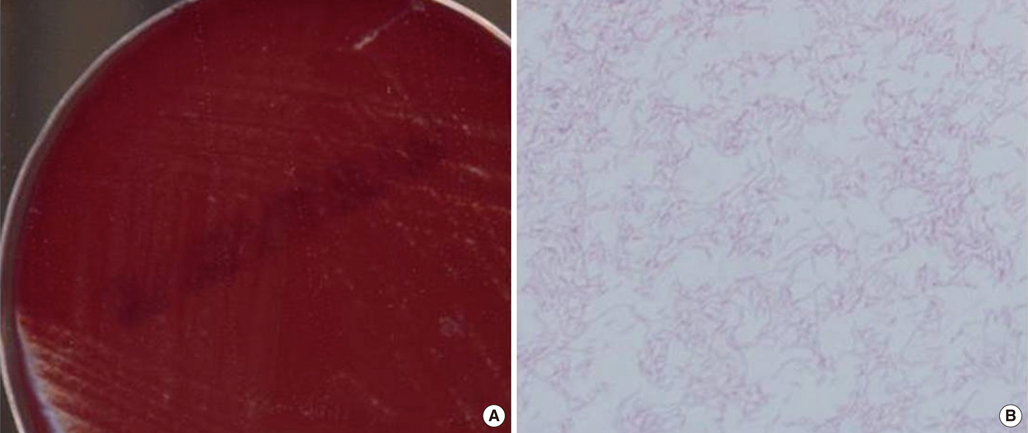
- Helicobacter cinaedi is an enterohepatic species. It can cause bacteremia, gastroenteritis, and cellulitis, particularly in immunocompromised individuals, such as those with acquired immunodeficiency syndrome, malignancy, or alcoholism. There are no previous reports of H. cinaedi infection in Korea. A 71-yr-old man was admitted to the emergency room because of dyspnea on November 9, 2011. He had undergone splenectomy 3 yr ago because of immune hemolytic anemia. Chest plain radiography revealed bilateral pleural effusion. He developed fever on hospital day (HD) 21. Three sets of blood cultures were taken, and gram-negative spiral bacilli were detected in all aerobic vials. The isolate grew in tiny colonies on chocolate agar after 3-day incubation under microaerophilic conditions. This organism tested positive for catalase and oxidase, and negative for urease. The 16S rRNA gene sequence of this isolate exhibited 99.8% homology with the published sequence of H. cinaedi CCUG 18818T (GenBank accession no. ABQT01000054) and 98.5% homology with the sequence of Helicobacter bilis Hb1T (GenBank accession no. U18766). The patient was empirically treated with piperacillin/tazobactam and levofloxacin, and discharged with improvement on HD 31. To our knowledge, this is the first report of H. cinaedi bacteremia in an asplenic patient. Asplenia appears to be a risk factor for H. cinaedi bacteremia.
Keyword
MeSH Terms
Figure
Reference
-
1. Gebhart CJ, Fennell CL, Murtaugh MP, Stamm WE. Campylobacter cinaedi is normal intestinal flora in hamsters. J Clin Microbiol. 1989. 27:1692–1694.2. Quinn TC, Goodell SE, Fennell C, Wang SP, Schuffler MD, Holmes KK, et al. Infections with Campylobacter jejuni and Campylobacter-like organisms in homosexual men. Ann Intern Med. 1984. 101:187–192.3. Matsumoto T, Goto M, Murakami H, Tanaka T, Nishiyama H, Ono E, et al. Multicenter study to evaluate bloodstream infection by Helicobacter cinaedi in Japan. J Clin Microbiol. 2007. 45:2853–2857.4. Holst H, Andresen K, Blom J, Højlyng N, Kemp M, Krogfelt KA, et al. A case of Helicobacter cinaedi bacteraemia in a previously healthy person with cellulitis. Open Microbiol J. 2008. 2:29–31.5. Relman DA. Persing DH, Smith TF, editors. Universal bacterial 16S rRNA amplification and sequencing. Diagnostic molecular microbiology principles and applications. 1993. 1st ed. Rochester, MN: Mayo Foundation;489–495.6. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007. 24:1596–1599.
Article7. Piccolomini R, Di Bonaventura G, Catamo G, Carbone F, Neri M. Comparative evaluation of the E test, agar dilution, and broth microdilution for testing susceptibilities of Helicobacter pylori strains to 20 antimicrobial agents. J Clin Microbiol. 1997. 35:1842–1846.
Article8. Clinical and Laboratory Standards Institute. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; Approved guideline-Second edition, M45-A2. 2010. Wayne, PA: Clinical and Laboratory Standards Institute.9. Minauchi K, Takahashi S, Sakai T, Kondo M, Shibayama K, Arakawa Y, et al. The nosocomial transmission of Helicobacter cinaedi infections in immunocompromised patients. Intern Med. 2010. 49:1733–1739.10. Taylor NS, Ge Z, Shen Z, Dewhirst FE, Fox JG. Cytolethal distending toxin: a potential virulence factor for Helicobacter cinaedi. J Infect Dis. 2003. 188:1892–1897.11. Solnick JV. Clinical significance of Helicobacter species other than Helicobacter pylori. Clin Infect Dis. 2003. 36:349–354.12. Sakran W, Raz R, Levi Y, Colodner R, Koren A. Campylobacter bacteremia and pneumonia in two splenectomized patients. Eur J Clin Microbiol Infect Dis. 1999. 18:496–498.13. Jackson N, Zaki M, Rahman AR, Nazim M, Win MN, Osman S. Fatal Campylobacter jejuni infection in a patient splenectomised for thalassaemia. J Clin Pathol. 1997. 50:436–437.14. Eraklis AJ, Filler RM. Splenectomy in childhood: a review of 1413 cases. J Pediatr Surg. 1972. 7:382–388.
Article15. Altamura M, Caradonna L, Amati L, Pellegrino NM, Urgesi G, Miniello S. Splenectomy and sepsis: the role of the spleen in the immune-mediated bacterial clearance. Immunopharmacol Immunotoxicol. 2001. 23:153–161.
Article16. Andersen LP. New Helicobacter species in humans. Dig Dis. 2001. 19:112–115.17. Van Genderen PJ, Goessens WH, Petit PL. Helicobacter cinaedi-associated bacteraemia and erysipelas in an immunocompetent host: a diagnostic challenge. Scand J Infect Dis. 2005. 37:382–385.18. Kiehlbauch JA, Brenner DJ, Cameron DN, Steigerwalt AG, Makowski JM, Baker CN, et al. Genotypic and phenotypic characterization of Helicobacter cinaedi and Helicobacter fennelliae strains isolated from humans and animals. J Clin Microbiol. 1995. 33:2940–2947.19. Lawson AJ. Versalovic J, Carroll KC, editors. Helicobacter. Manual of clinical microbiology. 2011. 10th ed. Washington, D.C.: ASM Press;900–915.
Article20. Clinical and Laboratory Standards Institute. Interpretive Criteria for Identification of Bacteria and Fungi by DNA Target Sequencing, Approved Guideline, MM18-A. 2008. Wayne, PA: Clinical and Laboratory Standards Institute.21. Vandamme P, Harrington CS, Jalava K, On SL. Misidentifying helicobacters: the Helicobacter cinaedi example. J Clin Microbiol. 2000. 38:2261–2266.22. Fernandez KR, Hansen LM, Vandamme P, Beaman BL, Solnick JV. Captive rhesus monkeys (Macaca mulatta) are commonly infected with Helicobacter cinaedi. J Clin Microbiol. 2002. 40:1908–1912.23. Kiehlbauch JA, Tauxe RV, Baker CN, Wachsmuth IK. Helicobacter cinaedi - associated bacteremia and cellulitis in immunocompromised patients. Ann Intern Med. 1994. 121:90–93.24. Orlicek SL, Welch DF, Kuhls TL. Septicemia and meningitis caused by Helicobacter cinaedi in a neonate. J Clin Microbiol. 1993. 31:569–571.25. Kitamura T, Kawamura Y, Ohkusu K, Masaki T, Iwashita H, Sawa T, et al. Helicobacter cinaedi cellulitis and bacteremia in immunocompetent hosts after orthopedic surgery. J Clin Microbiol. 2007. 45:31–38.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Gastric Ulcer Bleeding Caused by Systemic Inflammatory Response to Mycobacterial Bacteremia
- A Case of Shigella sonnei Bacteremia in an Adult
- Eggerthella Lenta Bacteremia after Appendectomy in a Healthy Patient
- Ignatzschineria larvae Bacteremia Following Lucilia sp. Myiasis in an Irregular Migrant: A Case Report
- A Case of Pyelonephritis Accompanied by Enterococcus hirae Bacteremia