Ann Lab Med.
2018 May;38(3):189-195. 10.3343/alm.2018.38.3.189.
Characteristics and Immunological Roles of Surface Layer Proteins in Clostridium difficile
- Affiliations
-
- 1Department of General Internal Medicine, National Hospital Organization Tokyo Medical Center, Meguro-ku, Tokyo, Japan. nobuaki.m@icloud.com
- 2Laboratory of Infectious Diseases, Graduate School of Infection Control Sciences and Kitasato Institute for Life Sciences, Kitasato University, Minato-ku, Tokyo, Japan.
- KMID: 2403451
- DOI: http://doi.org/10.3343/alm.2018.38.3.189
Abstract
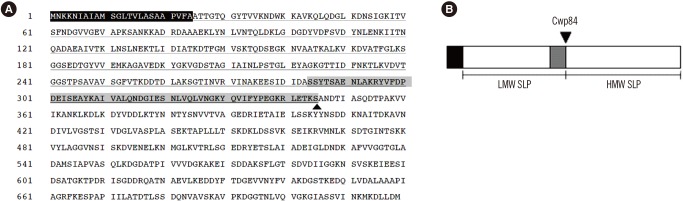
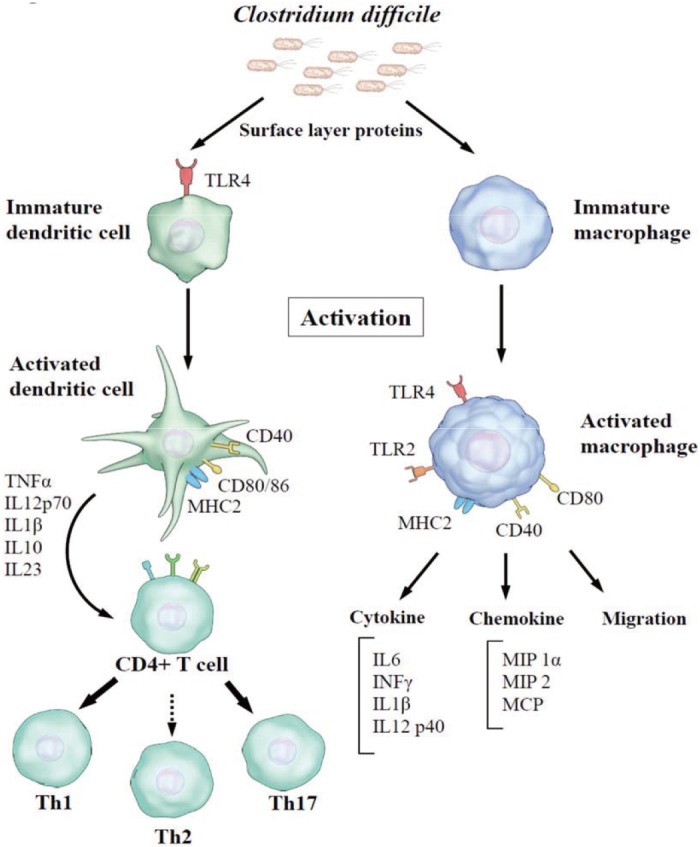
- Clostridium difficile is a major causative agent of antibiotic-associated diarrhea and has become the most common pathogen of healthcare-associated infection worldwide. The pathogenesis of C. difficile infection (CDI) is mediated by many factors such as colonization involving attachment to host intestinal epithelial cells, sporulation, germination, and toxin production. Bacterial cell surface components are crucial for the interaction between the bacterium and host cells. C. difficile has two distinct surface layer proteins (SLPs): a conserved high-molecular-weight SLP and a highly variable low-molecular-weight SLP. Recent studies have shown that C. difficile SLPs play roles not only in growth and survival, but also in adhesion to host epithelial cells and induction of cytokine production. Sequence typing of the variable region of the slpA gene, which encodes SLPs, is one of the methods currently used for typing C. difficile. SLPs have received much attention in recent years as vaccine candidates and new therapeutic agents in the treatment of C. difficile-associated diseases. Gaining mechanistic insights into the molecular functions of C. difficile SLPs will help advance our understanding of CDI pathogenesis and the development of vaccines and new therapeutic approaches. In this review, we summarize the characteristics and immunological roles of SLPs in C. difficile.
MeSH Terms
Figure
Reference
-
1. Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, et al. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994; 44:812–826. PMID: 7981107.2. Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013; 15:2631–2641. PMID: 23834245.3. Lawson PA, Citron DM, Tyrrell KL, Finegold SM. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prévot 1938. Anaerobe. 2016; 40:95–99. PMID: 27370902.4. Oren A, Garrity GM. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol. 2018; 68:1–2. PMID: 29292690.5. Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015; 372:825–834. PMID: 25714160.6. Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, et al. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev. 2010; 23:529–549. PMID: 20610822.7. Mori N, Aoki Y. Clinical characteristics and risk factors for community-acquired Clostridium difficile infection: a retrospective, case-control study in a tertiary care hospital in Japan. J Infect Chemother. 2015; 21:864–867. PMID: 26482373.8. Hensgens MP, Goorhuis A, Dekkers OM, van Benthem BHB, Kuijper EJ. All-cause and disease-specific mortality in hospitalized patients with Clostridium difficile infection: a multicenter cohort study. Clin Infect Dis. 2013; 56:1108–1116. PMID: 23300235.9. Honda H, Yamazaki A, Sato Y, Dubberke ER. Incidence and mortality associated with Clostridium difficile infection at a Japanese tertiary care center. Anaerobe. 2014; 25:5–10. PMID: 24184291.10. Takahashi M, Mori N, Bito S. Multi-institution case-control and cohort study of risk factors for the development and mortality of Clostridium difficile infections in Japan. BMJ Open. 2014; 4:e005665.11. Bloomfield MG, Sherwin JC, Gkrania-Klotsas E. Risk factors for mortality in Clostridium difficile infection in the general hospital population: a systematic review. J Hosp Infect. 2012; 82:1–12. PMID: 22727824.12. Collins DA, Hawkey PM, Riley TV. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control. 2013; 2:21. PMID: 23816346.13. Peniche AG, Savidge TC, Dann SM. Recent insights into Clostridium difficile pathogenesis. Curr Opin Infect Dis. 2013; 26:447–451. PMID: 23982235.14. Calabi E, Ward S, Wren B, Paxton T, Panico M, Morris H, et al. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol Microbiol. 2001; 40:1187–1199. PMID: 11401722.15. Fagan RP, Fairweather NF. Biogenesis and functions of bacterial Slayers. Nat Rev Microbiol. 2014; 12:211–222. PMID: 24509785.16. Masuda K, Itoh M, Kawata T. Characterization and reassembly of a regular array in the cell wall of Clostridium difficile GAI 4131. Microbiol Immunol. 1989; 33:287–298. PMID: 2770560.17. Ryan A, Lynch M, Smith SM, Amu S, Nel HJ, McCoy CE, et al. A role for TLR4 in Clostridium difficile infection and the recognition of surface layer proteins. PLoS Pathog. 2011; 7:e1002076. PMID: 21738466.18. Bianco M, Fedele G, Quattrini A, Spigaglia P, Barbanti F, Mastrantonio P, et al. Immunomodulatory activities of surface-layer proteins obtained from epidemic and hypervirulent Clostridium difficile strains. J Med Microbiol. 2011; 60:1162–1167. PMID: 21349985.19. Collins LE, Lynch M, Marszalowska I, Kristek M, Rochfort K, O'Connell M, et al. Surface layer proteins isolated from Clostridium difficile induce clearance responses in macrophages. Microbes Infect. 2014; 16:391–400. PMID: 24560642.20. Sára M, Sleytr UB. S-layer proteins. J Bacteriol. 2000; 182:859–868. PMID: 10648507.21. Fagan RP, Albesa-Jové D, Qazi O, Svergun DI, Brown KA, Fairweather NF. Structural insights into the molecular organization of the S-layer from Clostridium difficile. Mol Microbiol. 2009; 71:1308–1322. PMID: 19183279.22. Kirk JA, Banerji O, Fagan RP. Characteristics of the Clostridium difficile cell envelope and its importance in therapeutics. Microb Biotechnol. 2017; 10:76–90. PMID: 27311697.23. Qazi O, Hitchen P, Tissot B, Panico M, Morris HR, Dell A, et al. Mass spectrometric analysis of the S-layer proteins from Clostridium difficile demonstrates the absence of glycosylation. J Mass Spectrom. 2009; 44:368–374. PMID: 18932172.24. Sarker MR, Paredes-Sabja D. Molecular basis of early stages of Clostridium difficile infection: germination and colonization. Future Microbiol. 2012; 7:933–943. PMID: 22913353.25. Takeoka A, Takumi K, Koga T, Kawata T. Purification and characterization of S layer proteins from Clostridium difficile GAI 0714. J Gen Microbiol. 1991; 137:261–267. PMID: 1901902.26. Fagan RP, Fairweather NF. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem. 2011; 286:27483–27493. PMID: 21659510.27. Fagan RP, Janoir C, Collignon A, Mastrantonio P, Poxton IR, Fairweather NF. A proposed nomenclature for cell wall proteins of Clostridium difficile. J Med Microbiol. 2011; 60:1225–1228. PMID: 21252271.28. de la Riva L, Willing SE, Tate EW, Fairweather NF. Roles of cysteine proteases Cwp84 and Cwp13 in biogenesis of the cell wall of Clostridium difficile. J Bacteriol. 2011; 193:3276–3285. PMID: 21531808.29. Kandalaft H, Hussack G, Aubry A, van Faassen H, Guan Y, Arbabi-Ghahroudi M, et al. Targeting surface-layer proteins with single-domain antibodies: a potential therapeutic approach against Clostridium difficile-associated disease. Appl Microbiol Biotechnol. 2015; 99:8549–8562. PMID: 25936376.30. Karjalainen T, Waligora-Dupriet AJ, Cerquetti M, Spigaglia P, Maggioni A, Mauri P, et al. Molecular and genomic analysis of genes encoding surface-anchored proteins from Clostridium difficile. Infect Immun. 2001; 69:3442–3446. PMID: 11292772.31. Kato H, Yokoyama T, Arakawa Y. Typing by sequencing the slpA gene of Clostridium difficile strains causing multiple outbreaks in Japan. J Med Microbiol. 2005; 54:167–171. PMID: 15673512.32. Calabi E, Calabi F, Phillips AD, Fairweather NF. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect Immun. 2002; 70:5770–5778. PMID: 12228307.33. Dingle KE, Didelot X, Ansari MA, Eyre DW, Vaughan A, Griffiths D, et al. Recombinational switching of the Clostridium difficile S-layer and a novel glycosylation gene cluster revealed by large-scale whole-genome sequencing. J Infect Dis. 2013; 207:675–686. PMID: 23204167.34. Tasteyre A, Barc MC, Collignon A, Boureau H, Karjalainen T. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect Immun. 2001; 69:7937–7940. PMID: 11705981.35. Ternan NG, Jain S, Srivastava M, McMullan G. Comparative transcriptional analysis of clinically relevant heat stress response in Clostridium difficile strain 630. PLoS One. 2012; 7:e42410. PMID: 22860125.36. Waligora AJ, Hennequin C, Mullany P, Bourlioux P, Collignon A, Karjalainen T. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect Immun. 2001; 69:2144–2153. PMID: 11254569.37. Janoir C, Barc MC, Collignon A, Karjalainen T. Identification and characterization of a fibronectin-binding protein from Clostridium difficile. Microbiology. 2003; 149:2779–2787. PMID: 14523111.38. Cerquetti M, Molinari A, Sebastianelli A, Diociaiuti M, Petruzzelli R, Capo C, et al. Characterization of surface layer proteins from different Clostridium difficile clinical isolates. Microb Pathog. 2000; 28:363–372. PMID: 10839973.39. Gerding DN, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT. Gut Microbes. 2014; 5:15–27. PMID: 24253566.40. Merrigan MM, Venugopal A, Roxas JL, Anwar F, Mallozzi MJ, Roxas BA, et al. Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLoS One. 2013; 8:e78404. PMID: 24265687.41. Spigaglia P, Barketi-Klai A, Collignon A, Mastrantonio P, Barbanti F, Rupnik M, et al. Surface-layer (S-layer) of human and animal Clostridium difficile strains and their behaviour in adherence to epithelial cells and intestinal colonization. J Med Microbiol. 2013; 62:1386–1393. PMID: 23518658.42. Madan R, Petri WA Jr. Immune responses to Clostridium difficile infection. Trends Mol Med. 2012; 18:658–666. PMID: 23084763.43. Marsh JW, Arora R, Schlackman JL, Shutt KA, Curry SR, Harrison LH. Association of relapse of Clostridium difficile disease with BI/NAP1/027. J Clin Microbiol. 2012; 50:4078–4082. PMID: 23052318.44. Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, Notermans DW, et al. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis. 2008; 47:1162–1170. PMID: 18808358.45. Walk ST, Micic D, Jain R, Lo ES, Trivedi I, Liu EW, et al. Clostridium difficile ribotype does not predict severe infection. Clin Infect Dis. 2012; 55:1661–1668. PMID: 22972866.46. Lynch M, Walsh TA, Marszalowska I, Webb AE, MacAogain M, Rogers TR, et al. Surface layer proteins from virulent Clostridium difficile ribotypes exhibit signatures of positive selection with consequences for innate immune response. BMC Evol Biol. 2017; 17:135. PMID: 28606132.47. Knight DR, Elliott B, Chang BJ, Perkins TT, Riley TV. Diversity and evolution in the genome of Clostridium difficile. Clin Microbiol Rev. 2015; 28:721–741. PMID: 26085550.48. Karjalainen T, Saumier N, Barc MC, Delmée M, Collignon A. Clostridium difficile genotyping based on slpA variable region in S-layer gene sequence: an alternative to serotyping. J Clin Microbiol. 2002; 40:2452–2458. PMID: 12089261.49. McCoubrey J, Starr J, Martin H, Poxton IR. Clostridium difficile in a geriatric unit: a prospective epidemiological study employing a novel S-layer typing method. J Med Microbiol. 2003; 52:573–578. PMID: 12808079.50. Eidhin DN, Ryan AW, Doyle RM, Walsh JB, Kelleher D. Sequence and phylogenetic analysis of the gene for surface layer protein, slpA, from 14 PCR ribotypes of Clostridium difficile. J Med Microbiol. 2006; 55:69–83. PMID: 16388033.51. Poilane I, Humeniuk-Ainouz C, Durand I, Janoir C, Cruaud P, Delmée M, et al. Molecular characterization of Clostridium difficile clinical isolates in a geriatric hospital. J Med Microbiol. 2007; 56:386–390. PMID: 17314371.52. Kato H, Kato H, Nakamura M, Iwashima Y, Nakamura A, Ueda R, et al. Rapid analysis of Clostridium difficile strains recovered from hospitalized patients by using the slpA sequence typing system. J Infect Chemother. 2009; 15:199–202. PMID: 19554407.53. Joost I, Speck K, Herrmann M, von Müller L. Characterization of Clostridium difficile isolates by slpA and tcdC gene sequencing. Int J Antimicrob Agents. 2009; 33(Suppl 1):S13–S18. PMID: 19303562.54. Kato H, Kato H, Ito Y, Akahane T, Izumida S, Yokoyama T, et al. Typing of Clostridium difficile isolates endemic in Japan by sequencing of slpA and its application to direct typing. J Med Microbiol. 2010; 59:556–562. PMID: 20133413.55. Tagashira Y, Kato H, Senoh M, Nakamura A. Two cases of fulminant colitis due to binary toxin-positive Clostridium difficile that are not PCR ribotype 027 or type 078. J Med Microbiol. 2013; 62:1486–1489. PMID: 23558137.56. Xiao K, Kong F, Wang Q, Jin P, Thomas L, Xiong L, et al. Multiplex PCR targeting slpA: a rapid screening method to predict common Clostridium difficile ribotypes among fluoroquinolone resistant clinical strains. Pathology. 2013; 45:595–599. PMID: 24018815.57. Niwa H, Kato H, Hobo S, Kinoshita Y, Ueno T, Katayama Y, et al. Postoperative Clostridium difficile infection with PCR ribotype 078 strain identified at necropsy in five Thoroughbred racehorses. Vet Rec. 2013; 173:607.58. Stahlmann J, Schönberg M, Herrmann M, von Müller L. Detection of nosocomial Clostridium difficile infections with toxigenic strains despite negative toxin A and B testing on stool samples. Clin Microbiol Infect. 2014; 20:O590–O592. PMID: 24450741.59. von Müller L, Mock M, Halfmann A, Stahlmann J, Simon A, Herrmann M. Epidemiology of Clostridium difficile in Germany based on a single center long-term surveillance and German-wide genotyping of recent isolates provided to the advisory laboratory for diagnostic reasons. Int J Med Microbiol. 2015; 305:807–813. PMID: 26341328.60. Cheng JW, Xiao M, Kudinha T, Kong F, Xu ZP, Sun LY, et al. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolates from a university teaching hospital in China. Front Microbiol. 2016; 7:1621. PMID: 27799923.61. Miller-Roll T, Na’amnih W, Cohen D, Carmeli Y, Adler A. Molecular types and antimicrobial susceptibility patterns of Clostridium difficile isolates in different epidemiological settings in a tertiary care center in Israel. Diagn Microbiol Infect Dis. 2016; 86:450–454. PMID: 27638350.62. Killgore G, Thompson A, Johnson S, Brazier J, Kuijper E, Pepin J, et al. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J Clin Microbiol. 2008; 46:431–437. PMID: 18039796.63. Huber CA, Foster NF, Riley TV, Paterson DL. Challenges for standardization of Clostridium difficile typing methods. J Clin Microbiol. 2013; 51:2810–2814. PMID: 23784128.64. Drudy D, Calabi E, Kyne L, Sougioultzis S, Kelly E, Fairweather N, et al. Human antibody response to surface layer proteins in Clostridium difficile infection. FEMS Immunol Med Microbiol. 2004; 41:237–242. PMID: 15196573.65. Sánchez-Hurtado K, Corretge M, Mutlu E, McIlhagger R, Starr JM, Poxton IR. Systemic antibody response to Clostridium difficile in colonized patients with and without symptoms and matched controls. J Med Microbiol. 2008; 57:717–724. PMID: 18480328.66. Shirvan AN. Isolation of recombinant antibodies directed against surface proteins of Clostridium difficile. Braz J Microbiol. 2016; 47:394–402. PMID: 26991284.67. O'Brien JB, McCabe MS, Athié-Morales V, McDonald GS, Ní Eidhin DB, Kelleher DP. Passive immunization of hamsters against Clostridium difficile infection using antibodies to surface layer proteins. FEMS Microbiol Lett. 2005; 246:199–205. PMID: 15899406.68. Ní Eidhin DB, O'Brien JB, McCabe MS, Athié-Morales V, Kelleher DP. Active immunization of hamsters against Clostridium difficile infection using surface-layer protein. FEMS Immunol Med Microbiol. 2008; 52:207–218. PMID: 18093141.69. Bruxelle JF, Mizrahi A, Hoys S, Collignon A, Janoir C, Péchiné S. Immunogenic properties of the surface layer precursor of Clostridium difficile and vaccination assays in animal models. Anaerobe. 2016; 37:78–84. PMID: 26505926.70. Virdi V, Coddens A, De Buck S, Millet S, Goddeeris BM, Cox E, et al. Orally fed seeds producing designer IgAs protect weaned piglets against enterotoxigenic Escherichia coli infection. Proc Natl Acad Sci U S A. 2013; 110:11809–11814. PMID: 23801763.71. Siontorou CG. Nanobodies as novel agents for disease diagnosis and therapy. Int J Nanomedicine. 2013; 8:4215–4227. PMID: 24204148.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Characteristics of Clostridium difficile colitis
- SDS-PAGE profiles of clostridium difficile isolated from patientsand hospital environments
- New Treatment Option for Recurrent Clostridium difficile Infection
- Determination of toxin production of clostridium difficile strains isolated from patients with suspected antibiotic associated diarrhea
- Comparison of Clostridium difficile Toxin A Immunoassay with Cytotoxicity Assay