Blood Res.
2017 Mar;52(1):55-61. 10.5045/br.2017.52.1.55.
Characterization of clonal immunoglobulin heavy (IGH) V-D-J gene rearrangements and the complementarity-determining region in South Indian patients with precursor B-cell acute lymphoblastic leukemia
- Affiliations
-
- 1Department of Molecular Oncology, Cancer Institute (WIA), Chennai, India. nsudha79@gmail.com
- 2Department of Hematology and Immunology, Cancer Institute (WIA), Chennai, India.
- 3Department of Biotechnology, Dr. M.G.R. Educational & Research Institute, Chennai, India.
- KMID: 2375205
- DOI: http://doi.org/10.5045/br.2017.52.1.55
Abstract
- BACKGROUND
This study characterized clonal IG heavy V-D-J (IGH) gene rearrangements in South Indian patients with precursor B-cell acute lymphoblastic leukemia (precursor B-ALL) and identified age-related predominance in VDJ rearrangements.
METHODS
IGH rearrangements were studied in 50 precursor B-ALL cases (common ALL=37, pre-B ALL=10, pro-B ALL=3) by polymerase chain reaction (PCR) heteroduplex analysis. Twenty randomly selected clonal IGH rearrangement sequences were analyzed using the IMGT/V-QUEST tool.
RESULTS
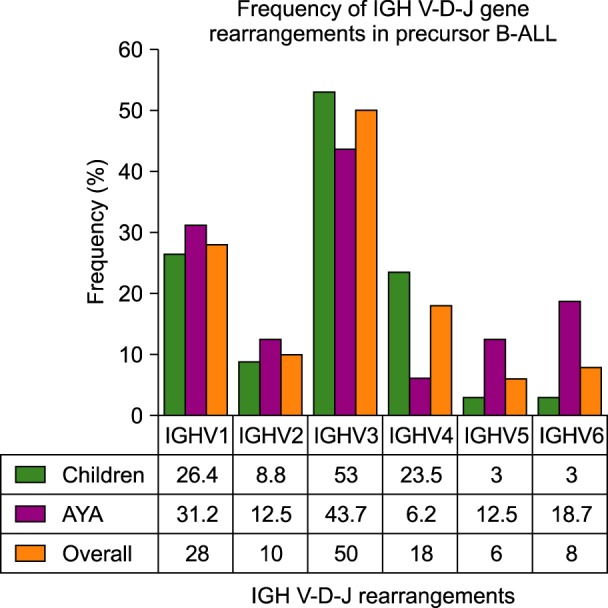
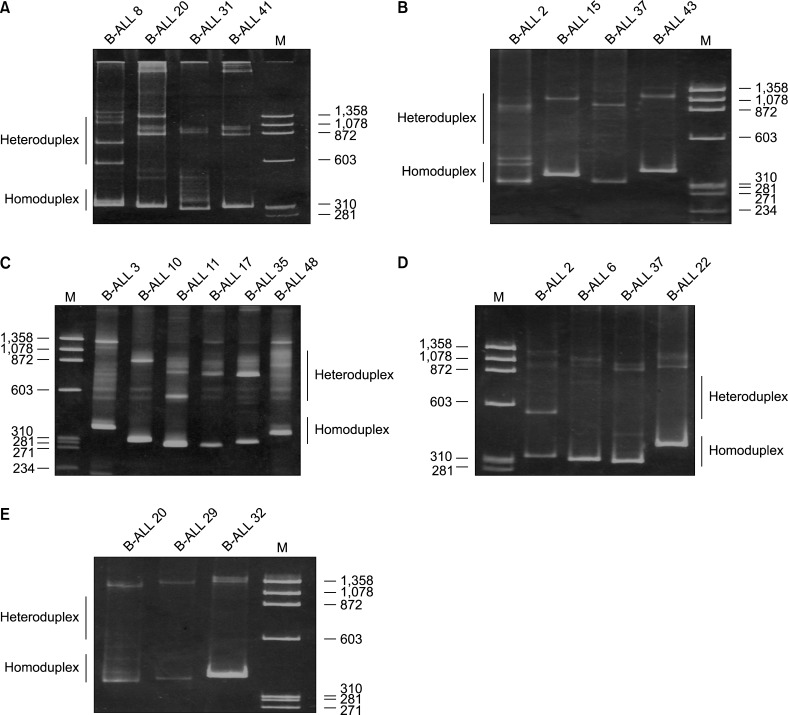
Clonal IGH rearrangements were detected in 41 (82%) precursor B-ALL cases. Among the IGHV1-IGHV7 subgroups, IGHV3 was used in 25 (50%) cases. Among the IGHD1-IGHD7 genes, IGHD2 and IGHD3 were used in 8 (40%) and 5 (25%) clones, respectively. Among the IGHJ1-IGHJ6 genes, IGHJ6 and IGHJ4 were used in 9 (45%) and 6 (30%) clones, respectively. In 6 out of 20 (30%) IGH rearranged sequences, CDR3 was in frame whereas 14 (70%) had rearranged sequences and CDR3 was out of frame. A somatic mutation in Vmut/Dmut/Jmut was detected in 14 of 20 IGH sequences. On average, Vmut/Dmut/Jmut were detected in 0.1 nt, 1.1 nt, and 0.2 nt, respectively.
CONCLUSION
The IGHV3 gene was frequently used whereas lower frequencies of IGHV5 and IGHV6 and a higher frequency of IGHV4 were detected in children compared with young adults. The IGHD2 and IGHD3 genes were over-represented, and the IGHJ6 gene was predominantly used in precursor-B-ALL. However, the IGH gene rearrangements in precursor-B-ALL did not show any significant age-associated genotype pattern attributed to our population.
MeSH Terms
Figure
Reference
-
1. Tonegawa S. Somatic generation of antibody diversity. Nature. 1983; 302:575–581. PMID: 6300689.
Article2. Stankovic T, Weston V, McConville CM, et al. Clonal diversity of Ig and T-cell receptor gene rearrangements in childhood B-precursor acute lymphoblastic leukaemia. Leuk Lymphoma. 2000; 36:213–224. PMID: 10674894.
Article3. Matsuda F, Ishii K, Bourvagnet P, et al. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med. 1998; 188:2151–2162. PMID: 9841928.
Article4. Kabat EA. Antibody complementarity and antibody structure. J Immunol. 1988; 141:S25–S36. PMID: 3049810.5. Lefranc MP, Lefranc G. The immunoglobulin factsbook. London, UK: Academic Press;2001. p. 1–458.6. Giudicelli V, Lefranc MP. IMGT/junctionanalysis: IMGT standardized analysis of the V-J and V-D-J junctions of the rearranged immunoglobulins (IG) and T cell receptors (TR). Cold Spring Harb Protoc. 2011; 2011:716–725. PMID: 21632777.
Article7. Alt FW, Yancopoulos GD, Blackwell TK, et al. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984; 3:1209–1219. PMID: 6086308.
Article8. Sudhakar N, Nancy NK, Rajalekshmy KR, Ramanan G, Rajkumar T. T-cell receptor gamma and delta gene rearrangements and junctional region characteristics in south Indian patients with T-cell acute lymphoblastic leukemia. Am J Hematol. 2007; 82:215–221. PMID: 17133429.
Article9. Magrath I, Shanta V, Advani S, et al. Treatment of acute lymphoblastic leukaemia in countries with limited resources; lessons from use of a single protocol in India over a twenty year period. Eur J Cancer. 2005; 41:1570–1583. PMID: 16026693.10. Szczepański T, Willemse MJ, van Wering ER, van Weerden JF, Kamps WA, van Dongen JJ. Precursor-B-ALL with D(H)-J(H) gene rearrangements have an immature immunogenotype with a high frequency of oligoclonality and hyperdiploidy of chromosome 14. Leukemia. 2001; 15:1415–1423. PMID: 11516102.
Article11. Langerak AW, Szczepański T, van der Burg M, Wolvers-Tettero IL, van Dongen JJ. Heteroduplex PCR analysis of rearranged T cell receptor genes for clonality assessment in suspect T cell proliferations. Leukemia. 1997; 11:2192–2199. PMID: 9447840.
Article12. Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008; 36:W503–W508. PMID: 18503082.
Article13. Sazawal S, Bhatia K, Gurbuxani S, et al. Pattern of immunoglobulin (Ig) and T-cell receptor (TCR) gene rearrangements in childhood acute lymphoblastic leukemia in India. Leuk Res. 2000; 24:575–582. PMID: 10867131.
Article14. Li A, Rue M, Zhou J, et al. Utilization of Ig heavy chain variable, diversity, and joining gene segments in children with B-lineage acute lymphoblastic leukemia: implications for the mechanisms of VDJ recombination and for pathogenesis. Blood. 2004; 103:4602–4609. PMID: 15010366.
Article15. Schroeder HW Jr, Wang JY. Preferential utilization of conserved immunoglobulin heavy chain variable gene segments during human fetal life. Proc Natl Acad Sci U S A. 1990; 87:6146–6150. PMID: 2117273.
Article16. Yamada M, Wasserman R, Reichard BA, Shane S, Caton AJ, Rovera G. Preferential utilization of specific immunoglobulin heavy chain diversity and joining segments in adult human peripheral blood B lymphocytes. J Exp Med. 1991; 173:395–407. PMID: 1899102.
Article17. Wasserman R, Ito Y, Galili N, et al. The pattern of joining (JH) gene usage in the human IgH chain is established predominantly at the B precursor cell stage. J Immunol. 1992; 149:511–516. PMID: 1624797.18. Schroeder HW Jr, Mortari F, Shiokawa S, Kirkham PM, Elgavish RA, Bertrand FE 3rd. Developmental regulation of the human antibody repertoire. Ann N Y Acad Sci. 1995; 764:242–260. PMID: 7486531.19. Beishuizen A, Hahlen K, Hagemeijer A, et al. Multiple rearranged immunoglobulin genes in childhood acute lymphoblastic leukemia of precursor B-cell origin. Leukemia. 1991; 5:657–667. PMID: 1909409.20. Wasserman R, Yamada M, Ito Y, et al. VH gene rearrangement events can modify the immunoglobulin heavy chain during progression of B-lineage acute lymphoblastic leukemia. Blood. 1992; 79:223–228. PMID: 1728310.21. Steenbergen EJ, Verhagen OJ, van Leeuwen EF, von dem Borne AE, van der Schoot CE. Distinct ongoing Ig heavy chain rearrangement processes in childhood B-precursor acute lymphoblastic leukemia. Blood. 1993; 82:581–589. PMID: 8329713.22. Katsibardi K, Braoudaki M, Papathanasiou C, Karamolegou K, Tzortzatou-Stathopoulou F. Clinical significance of productive immunoglobulin heavy chain gene rearrangements in childhood acute lymphoblastic leukemia. Leuk Lymphoma. 2011; 52:1751–1757. PMID: 21649543.
Article23. Levy S, Mendel E, Kon S, Avnur Z, Levy R. Mutational hot spots in Ig V region genes of human follicular lymphomas. J Exp Med. 1988; 168:475–489. PMID: 3045247.
Article24. Kiyoi H, Naoe T, Horibe K, Ohno R. Characterization of the immunoglobulin heavy chain complementarity determining region (CDR)-III sequences from human B cell precursor acute lymphoblastic leukemia cells. J Clin Invest. 1992; 89:739–746. PMID: 1541668.
Article25. Gawad C, Pepin F, Carlton VE, et al. Massive evolution of the immunoglobulin heavy chain locus in children with B precursor acute lymphoblastic leukemia. Blood. 2012; 120:4407–4417. PMID: 22932801.
Article26. Li S, Lefranc MP, Miles JJ, et al. IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat Commun. 2013; 4:2333. PMID: 23995877.
Article27. Gazzola A, Mannu C, Rossi M, et al. The evolution of clonality testing in the diagnosis and monitoring of hematological malignancies. Ther Adv Hematol. 2014; 5:35–47. PMID: 24688753.
Article28. Sudhakar N, Nancy NK, Rajalekshmy KR, Rajkumar T. Diversity of T-cell receptor gene rearrangements in South Indian patients with common acute lymphoblastic leukemia. Iran J Immunol. 2009; 6:141–146. PMID: 19801787.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Submicroscopic Deletions of Immunoglobulin Heavy Chain Gene (IGH) in Precursor B Lymphoblastic Leukemia with IGH Rearrangements
- Non-Radioactive Detection of Clonality in Malignant Lymphoid Neoplasms using the Polymerase Chain Reaction
- Polymerase Chain Reaction and Sequencing of Immunoglobulin Heavy Chain Gene Rearrangement in Formalin Fixed, Paraffin-embedded Tissue of Patients with B Cell Lymphoma
- Precursor B-Cell Acute Lymphoblastic Leukemia in Two Patients with a History of Cytotoxic Therapy
- A Case of B-cell Precursor Acute Lymphoblastic Leukemia with the t(14;22)(q32;q11) Presenting Hyperleukocytosis