J Gastric Cancer.
2012 Jun;12(2):73-80.
Amplification of the UQCRFS1 Gene in Gastric Cancers
- Affiliations
-
- 1Department of General Surgery, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- 2Department of Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea. wonsang@catholic.ac.kr
Abstract
- PURPOSE
The specific aim of this study is to unravel a DNA copy number alterations, and to search for novel genes that are associated with the development of Korean gastric cancer.
MATERIALS AND METHODS
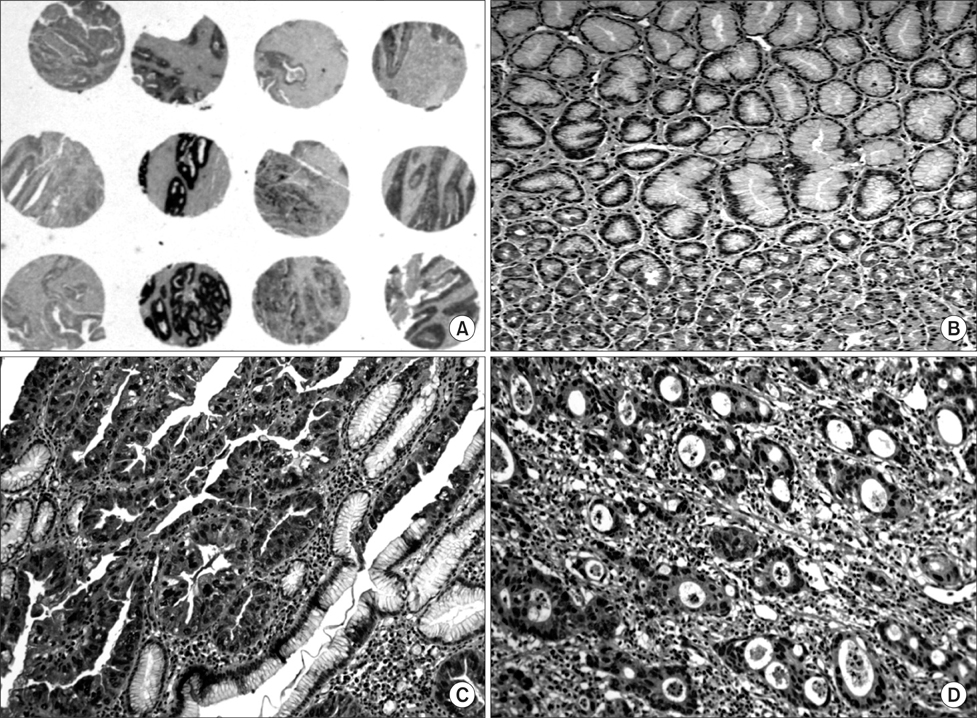
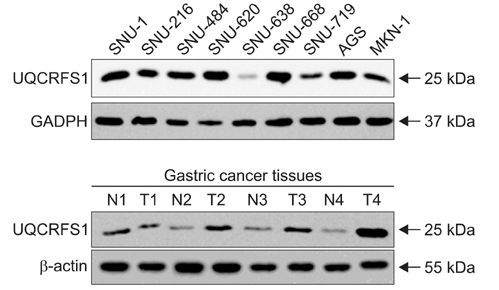
We investigated a DNA copy number changes in 23 gastric adenocarcinomas by array-comparative genomic hybridization and quantitative real-time polymerase chain reaction analyses. Besides, the expression of UQCRFS1, which shows amplification in array-CGH, was examined in 186 gastric cancer tissues by an immunohistochemistry, and in 9 gastric cancer cell lines, as well as 24 gastric cancer tissues by immunoblotting.
RESULTS
We found common gains at 48 different loci, and a common loss at 19 different loci. Amplification of UQCRFS1 gene at 19q12 was found in 5 (21.7%) of the 23 gastric cancers in an array-comparative genomic hybridization and DNA copy number were increased in 5 (20.0%) out of the 25 gastric cancer in quantitative real-time polymerase chain reaction. In immunohistochemistry, the overexpression of the protein was detected in 105 (56.5%) out of the 186 gastric cancer tissues. Statistically, there was no significant relationship between the overexpression of UQCRFS1 and clinicopathologic parameters (P>0.05). In parallel, the overexpression of UQCRFS1 protein was confirmed in 6 (66.7%) of the 9 gastric cancer cell lines, and 12 (50.0%) of the 24 gastric cancer tissues by immunoblotting.
CONCLUSIONS
These results suggest that the overexpression of UQCRFS1 gene may contribute to the development and/or progression of gastric cancer, and further supported that mitochondrial change may serve as a potential cancer biomarker.
MeSH Terms
Figure
Reference
-
1. Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005. 55:10–30.
Article2. Jung KW, Park S, Kong HJ, Won YJ, Lee JY, Park EC, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2008. Cancer Res Treat. 2011. 43:1–11.
Article3. Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001. 29:263–264.
Article4. Buffart TE, Carvalho B, Hopmans E, Brehm V, Kranenbarg EK, Schaaij-Visser TB, et al. Gastric cancers in young and elderly patients show different genomic profiles. J Pathol. 2007. 211:45–51.
Article5. Weiss MM, Kuipers EJ, Postma C, Snijders AM, Pinkel D, Meuwissen SG, et al. Genomic alterations in primary gastric adenocarcinomas correlate with clinicopathological characteristics and survival. Cell Oncol. 2004. 26:307–317.
Article6. Weiss MM, Kuipers EJ, Postma C, Snijders AM, Siccama I, Pinkel D, et al. Genomic profiling of gastric cancer predicts lymph node status and survival. Oncogene. 2003. 22:1872–1879.
Article7. Sugihara H, Hattori T, Fujita S, Hirose K, Fukuda M. Regional ploidy variations in signet ring cell carcinomas of the stomach. Cancer. 1990. 65:122–129.
Article8. Hattori T, Sugihara H, Fukuda M, Hamada S, Fujita S. DNA ploidy patterns of minute carcinomas in the stomach. Jpn J Cancer Res. 1986. 77:276–281.9. Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000. 132:365–386.
Article10. Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998. 4:844–847.
Article11. Yoon JH, Song JH, Zhang C, Jin M, Kang YH, Nam SW, et al. Inactivation of the Gastrokine 1 gene in gastric adenomas and carcinomas. J Pathol. 2011. 223:618–625.
Article12. Tsukamoto Y, Uchida T, Karnan S, Noguchi T, Nguyen LT, Tanigawa M, et al. Genome-wide analysis of DNA copy number alterations and gene expression in gastric cancer. J Pathol. 2008. 216:471–482.
Article13. Rossi E, Klersy C, Manca R, Zuffardi O, Solcia E. Correlation between genomic alterations assessed by array comparative genomic hybridization, prognostically informative histologic subtype, stage, and patient survival in gastric cancer. Hum Pathol. 2011. 42:1937–1945.
Article14. Bizari L, Borim AA, Leite KR, Gonçalves Fde T, Cury PM, Tajara EH, et al. Alterations of the CCND1 and HER-2/neu (ERBB2) proteins in esophageal and gastric cancers. Cancer Genet Cytogenet. 2006. 165:41–50.
Article15. Calcagno DQ, Leal MF, Seabra AD, Khayat AS, Chen ES, Demachki S, et al. Interrelationship between chromosome 8 aneuploidy, C-MYC amplification and increased expression in individuals from northern Brazil with gastric adenocarcinoma. World J Gastroenterol. 2006. 12:6207–6211.
Article16. Wolf M, Mousses S, Hautaniemi S, Karhu R, Huusko P, Allinen M, et al. High-resolution analysis of gene copy number alterations in human prostate cancer using CGH on cDNA microarrays: impact of copy number on gene expression. Neoplasia. 2004. 6:240–247.
Article17. Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002. 62:6240–6245.18. Järvinen AK, Autio R, Haapa-Paananen S, Wolf M, Saarela M, Grénman R, et al. Identification of target genes in laryngeal squamous cell carcinoma by high-resolution copy number and gene expression microarray analyses. Oncogene. 2006. 25:6997–7008.
Article19. Kim HK, Park WS, Kang SH, Warda M, Kim N, Ko JH, et al. Mitochondrial alterations in human gastric carcinoma cell line. Am J Physiol Cell Physiol. 2007. 293:C761–C771.
Article20. Ohashi Y, Kaneko SJ, Cupples TE, Young SR. Ubiquinol cytochrome c reductase (UQCRFS1) gene amplification in primary breast cancer core biopsy samples. Gynecol Oncol. 2004. 93:54–58.
Article21. Sait SN, Qadir MU, Conroy JM, Matsui S, Nowak NJ, Baer MR. Double minute chromosomes in acute myeloid leukemia and myelodysplastic syndrome: identification of new amplification regions by fluorescence in situ hybridization and spectral karyotyping. Genes Chromosomes Cancer. 2002. 34:42–47.
Article22. Kaneko SJ, Gerasimova T, Smith ST, Lloyd KO, Suzumori K, Young SR. CA125 and UQCRFS1 FISH studies of ovarian carcinoma. Gynecol Oncol. 2003. 90:29–36.
Article23. Rieske JS. Composition, structure, and function of complex III of the respiratory chain. Biochim Biophys Acta. 1976. 456:195–247.
Article24. Trumpower BL, Edwards CA. Purification of a reconstitutively active iron-sulfur protein (oxidation factor) from succinate. cytochrome c reductase complex of bovine heart mitochondria. J Biol Chem. 1979. 254:8697–8706.
Article25. Fiskum G, Starkov A, Polster BM, Chinopoulos C. Mitochondrial mechanisms of neural cell death and neuroprotective interventions in Parkinson's disease. Ann N Y Acad Sci. 2003. 991:111–119.
Article26. Owens KM, Kulawiec M, Desouki MM, Vanniarajan A, Singh KK. Impaired OXPHOS complex III in breast cancer. PLoS One. 2011. 6:e23846.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Amplification and Overexpression of c-erbB-2 in Gastric Cancer
- HER2 Status and Its Heterogeneity in Gastric Carcinoma of Vietnamese Patient
- C-met and E-cadherin Expression in Advanced Gastric Cancer
- HER2 Status in Gastric Adenocarcinomas Assessed by Immunohistochemistry, Automated Silver-Enhanced In Situ Hybridization and Fluorescence In Situ Hybridization
- Prevalence and Clinicopathological Significance of METOverexpression and Gene Amplification in Patients withGallbladder Carcinoma