Cancer Res Treat.
2020 Apr;52(2):481-491. 10.4143/crt.2019.370.
Prevalence and Clinicopathological Significance of METOverexpression and Gene Amplification in Patients withGallbladder Carcinoma
- Affiliations
-
- 1Department of Pathology, Hanyang University College of Medicine, Seoul, Korea
- KMID: 2500333
- DOI: http://doi.org/10.4143/crt.2019.370
Abstract
- Purpose
Mesenchymal epithelial transition (MET) is a proto-oncogene that encodes a heterodimeric transmembrane receptor tyrosine kinase for the hepatocyte growth factor. Aberrant MET signaling has been described in several solid tumors—especially non-small cell lung cancer— and is associated with tumor progression and adverse prognosis. As MET is a potential therapeutic target, information regarding its prevalence and clinicopathological relevance is crucial.
Materials and Methods
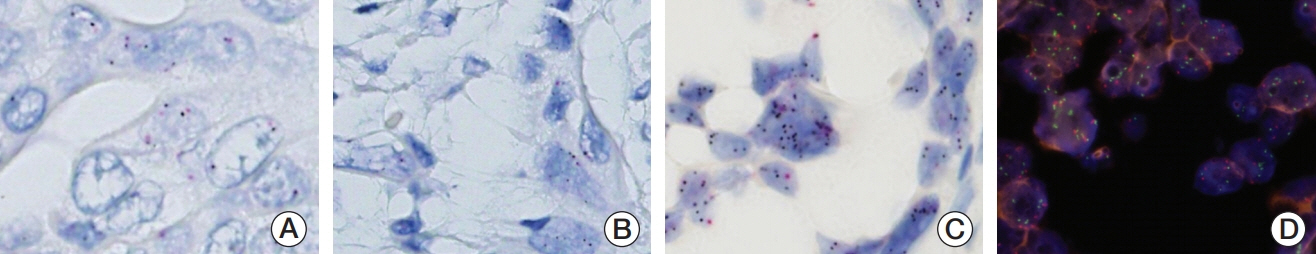
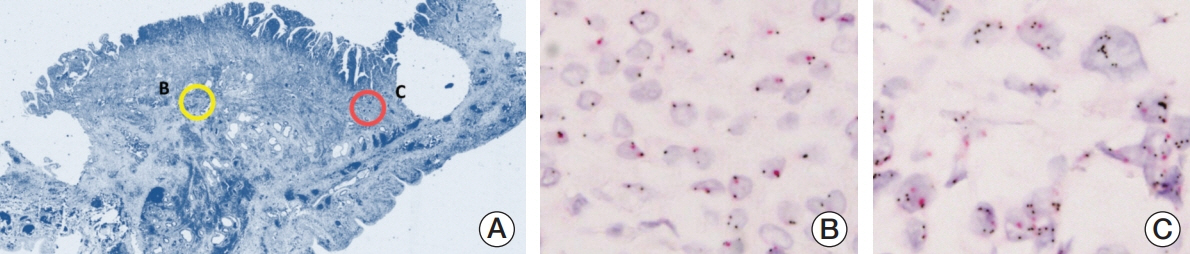
We investigated MET expression and gene amplification in 113 gallbladder cancers using tissue microarray. Immunohistochemistry was used to evaluate MET overexpression, and silver/fluorescence in situ hybridization (ISH) was used to assess gene copy number.
Results
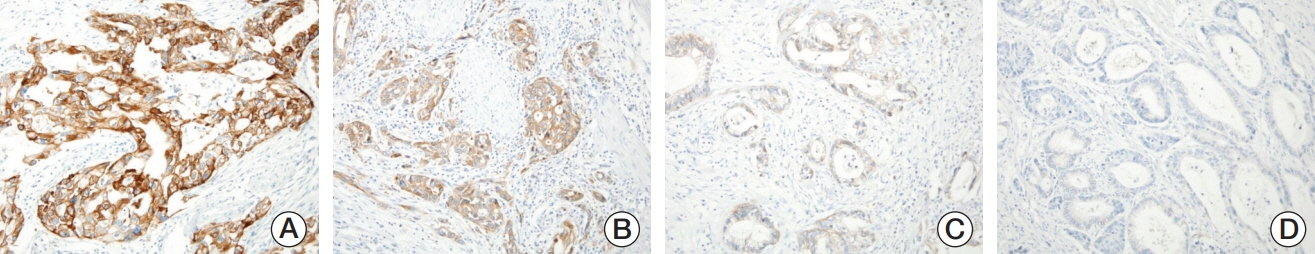
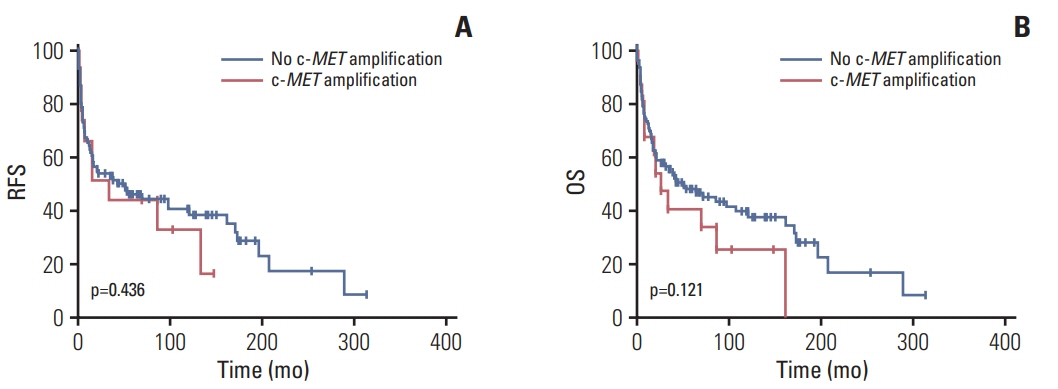
MET overexpression was found in 37 cases of gallbladder carcinoma (39.8%), and gene amplification was present in 17 cases (18.3%). MET protein expression did not correlate with MET amplification. MET amplification was significantly associated with aggressive clinicopathological features, including high histological grade, advanced pT category, lymph node metastasis, and advanced American Joint Committee on Cancer stage. There was no significant correlation between any clinicopathological factors and MET overexpression. No difference in survival was found with respect to MET overexpression and amplification status.
Conclusion
Our data suggested that MET might be a potential therapeutic target for targeted therapy in gallbladder cancer, because MET amplification was found in a subset of tumors associated with adverse prognostic factors. Detection of MET amplification by ISH might be a useful predictive biomarker test for anti-MET therapy.
Keyword
Figure
Reference
-
References
1. Jung KW, Won YJ, Kong HJ, Lee ES. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2016. Cancer Res Treat. 2019; 51:417–30.
Article2. Sharma A, Sharma KL, Gupta A, Yadav A, Kumar A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: recent update. World J Gastroenterol. 2017; 23:3978–98.
Article3. Li M, Zhang Z, Li X, Ye J, Wu X, Tan Z, et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet. 2014; 46:872–6.
Article4. Bengala C, Bertolini F, Malavasi N, Boni C, Aitini E, Dealis C, et al. Sorafenib in patients with advanced biliary tract carcinoma: a phase II trial. Br J Cancer. 2010; 102:68–72.
Article5. Chiorean EG, Ramasubbaiah R, Yu M, Picus J, Bufill JA, Tong Y, et al. Phase II trial of erlotinib and docetaxel in advanced and refractory hepatocellular and biliary cancers: Hoosier Oncology Group GI06-101. Oncologist. 2012; 17:13.
Article6. Vigna E, Naldini L, Tamagnone L, Longati P, Bardelli A, Maina F, et al. Hepatocyte growth factor and its receptor, the tyrosine kinase encoded by the c-MET proto-oncogene. Cell Mol Biol (Noisy-le-grand). 1994; 40:597–604.7. Blumenschein GR Jr, Mills GB, Gonzalez-Angulo AM. Targeting the hepatocyte growth factor-cMET axis in cancer therapy. J Clin Oncol. 2012; 30:3287–96.8. Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003; 4:915–25.
Article9. Furge KA, Zhang YW, Vande Woude GF. Met receptor tyrosine kinase: enhanced signaling through adapter proteins. Oncogene. 2000; 19:5582–9.
Article10. Nakamura Y, Niki T, Goto A, Morikawa T, Miyazawa K, Nakajima J, et al. c-Met activation in lung adenocarcinoma tissues: an immunohistochemical analysis. Cancer Sci. 2007; 98:1006–13.
Article11. Suzuki K, Hayashi N, Yamada Y, Yoshihara H, Miyamoto Y, Ito Y, et al. Expression of the c-met protooncogene in human hepatocellular carcinoma. Hepatology. 1994; 20:1231–6.
Article12. Drilon A, Cappuzzo F, Ou SI, Camidge DR. Targeting MET in lung cancer: will expectations finally be MET? J Thorac Oncol. 2017; 12:15–26.
Article13. Spigel DR, Ervin TJ, Ramlau RA, Daniel DB, Goldschmidt JH Jr, Blumenschein GR Jr, et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2013; 31:4105–14.
Article14. Schildhaus HU, Heukamp LC, Merkelbach-Bruse S, Riesner K, Schmitz K, Binot E, et al. Definition of a fluorescence in-situ hybridization score identifies high- and low-level FGFR1 amplification types in squamous cell lung cancer. Mod Pathol. 2012; 25:1473–80.
Article15. Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009; 27:1667–74.16. Tanaka A, Sueoka-Aragane N, Nakamura T, Takeda Y, Mitsuoka M, Yamasaki F, et al. Co-existence of positive MET FISH status with EGFR mutations signifies poor prognosis in lung adenocarcinoma patients. Lung Cancer. 2012; 75:89–94.
Article17. Jurmeister P, Lenze D, Berg E, Mende S, Schaper F, Kellner U, et al. Parallel screening for ALK, MET and ROS1 alterations in non-small cell lung cancer with implications for daily routine testing. Lung Cancer. 2015; 87:122–9.
Article18. Ou SH, Kwak EL, Siwak-Tapp C, Dy J, Bergethon K, Clark JW, et al. Activity of crizotinib (PF02341066), a dual mesenchymalepithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol. 2011; 6:942–6.
Article19. Sierra JR, Tsao MS. c-MET as a potential therapeutic target and biomarker in cancer. Ther Adv Med Oncol. 2011; 3(1 Suppl):S21–35.
Article20. Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984; 311:29–33.
Article21. Li Y, Chen CQ, He YL, Cai SR, Yang DJ, He WL, et al. Abnormal expression of E-cadherin in tumor cells is associated with poor prognosis of gastric carcinoma. J Surg Oncol. 2012; 106:304–10.
Article22. Di Renzo MF, Olivero M, Giacomini A, Porte H, Chastre E, Mirossay L, et al. Overexpression and amplification of the met/HGF receptor gene during the progression of colorectal cancer. Clin Cancer Res. 1995; 1:147–54.23. Garcia S, Dales JP, Jacquemier J, Charafe-Jauffret E, Birnbaum D, Andrac-Meyer L, et al. c-Met overexpression in inflammatory breast carcinomas: automated quantification on tissue microarrays. Br J Cancer. 2007; 96:329–35.
Article24. Radaeva S, Ferreira-Gonzalez A, Sirica AE. Overexpression of C-NEU and C-MET during rat liver cholangiocarcinogenesis: a link between biliary intestinal metaplasia and mucin-producing cholangiocarcinoma. Hepatology. 1999; 29:1453–62.
Article25. Nakazawa K, Dobashi Y, Suzuki S, Fujii H, Takeda Y, Ooi A. Amplification and overexpression of c-erbB-2, epidermal growth factor receptor, and c-met in biliary tract cancers. J Pathol. 2005; 206:356–65.26. Yang L, Guo T, Jiang S, Yang Z. Expression of ezrin, HGF and c-met and its clinicopathological significance in the benign and malignant lesions of the gallbladder. Hepatogastroenterology. 2012; 59:1769–75.27. Sanada Y, Osada S, Tokuyama Y, Tanaka Y, Takahashi T, Yamaguchi K, et al. Critical role of c-Met and Ki67 in progress of biliary carcinoma. Am Surg. 2010; 76:372–9.
Article28. Moon WS, Park HS, Lee H, Pai R, Tarnawski AS, Kim KR, et al. Co-expression of cox-2, C-met and beta-catenin in cells forming invasive front of gallbladder cancer. Cancer Res Treat. 2005; 37:171–6.29. Heo MH, Kim HK, Lee H, Kim KM, Lee J, Park SH, et al. The clinical impact of c-MET over-Expression in advanced biliary tract cancer (BTC). J Cancer. 2017; 8:1395–9.
Article30. Narayan RR, Creasy JM, Goldman DA, Gonen M, Kandoth C, Kundra R, et al. Regional differences in gallbladder cancer pathogenesis: Insights from a multi-institutional comparison of tumor mutations. Cancer. 2019; 125:575–85.
Article31. Lee H, Ross JS. The potential role of comprehensive genomic profiling to guide targeted therapy for patients with biliary cancer. Therap Adv Gastroenterol. 2017; 10:507–20.
Article32. Choi J, Lee HE, Lee HS, Han N, Kim MA, Kim WH. Evaluation of intratumoral and intertumoral heterogeneity of MET protein expression in gastric cancer. Appl Immunohistochem Mol Morphol. 2018; 26:445–53.
Article33. Casadevall D, Gimeno J, Clave S, Taus A, Pijuan L, Arumi M, et al. MET expression and copy number heterogeneity in nonsquamous non-small cell lung cancer (nsNSCLC). Oncotarget. 2015; 6:16215–26.
Article34. Lee SJ, Lee J, Sohn I, Mao M, Kai W, Park CK, et al. A survey of c-MET expression and amplification in 287 patients with hepatocellular carcinoma. Anticancer Res. 2013; 33:5179–86.35. Sun W, Song L, Ai T, Zhang Y, Gao Y, Cui J. Prognostic value of MET, cyclin D1 and MET gene copy number in non-small cell lung cancer. J Biomed Res. 2013; 27:220–30.
Article36. Appleman LJ. MET signaling pathway: a rational target for cancer therapy. J Clin Oncol. 2011; 29:4837–8.
Article37. Barat S, Bozko P, Chen X, Scholta T, Hanert F, Gotze J, et al. Targeting c-MET by LY2801653 for treatment of cholangiocarcinoma. Mol Carcinog. 2016; 55:2037–50.
Article38. Duan Z, Choy E, Nielsen GP, Rosenberg A, Iafrate J, Yang C, et al. Differential expression of microRNA (miRNA) in chordoma reveals a role for miRNA-1 in Met expression. J Orthop Res. 2010; 28:746–52.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- C-met and E-cadherin Expression in Advanced Gastric Cancer
- Clinicopathologic Analysis of Epidermal Growth Factor Receptor Status in Non-small Cell Lung Cancer: Protein Expression, Gene Amplification and Survival Analysis
- Amplification and Overexpression of c-erbB-2 in Gastric Cancer
- Correlation between Amplification of c-myc Oncogene and Histophthologic Prognostic Factors in Endomentrial Cancer
- Amplification and Overexpression of Cyclin D1 in Head and Neck Squamous Cell Carcinomas