J Bone Metab.
2016 Aug;23(3):121-127. 10.11005/jbm.2016.23.3.121.
Current Status and Strategy of microRNA Research for Cartilage Development and Osteoarthritis Pathogenesis
- Affiliations
-
- 1Department of Molecular and Experimental Medicine, The Scripps Research Institute, La Jolla, California, USA. asahara.syst@tmd.ac.jp
- 2Department of Systems BioMedicine, Tokyo Medical and Dental University, Tokyo, Japan.
- 3The Core Research for the Evolutionary Science and Technology from Japan Agency for Medical Research and Development, Tokyo, Japan.
- KMID: 2350807
- DOI: http://doi.org/10.11005/jbm.2016.23.3.121
Abstract
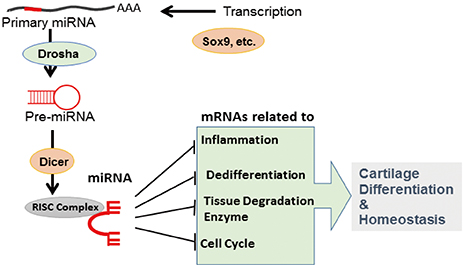
- MicroRNAs (miRNAs), which are small (~21 nucleotides) non-coding RNAs, are important players in endochondral ossification, articular cartilage homeostasis, and arthritis pathogenesis. Comprehensive and genetic analyses of cartilage-specific or cartilage-related miRNAs have provided new information on cartilage development, homeostasis, and related diseases. State-of-the-art combinatorial approaches, including transcription-activator like effector nuclease (TALEN)/clustered regularly interspaced short palindromic repeats (CRISPR) technique for targeting miRNAs and high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation for identifying target messenger RNAs, should be used to determine complex miRNA networks and miRNA-dependent cartilage regulation. Use of advanced drug delivery systems involving cartilage-specific miRNAs will accelerate the application of these new findings in arthritis therapy.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Regulation of Cartilage Development and Diseases by Transcription Factors
Riko Nishimura, Kenji Hata, Yoshifumi Takahata, Tomohiko Murakami, Eriko Nakamura, Hiroko Yagi
J Bone Metab. 2017;24(3):147-153. doi: 10.11005/jbm.2017.24.3.147.
Reference
-
1. Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997; 88:637–646.
Article2. Euling S, Ambros V. Heterochronic genes control cell cycle progress and developmental competence of C. elegans vulva precursor cells. Cell. 1996; 84:667–676.
Article3. Cancedda R, Descalzi Cancedda F, Castagnola P. Chondrocyte differentiation. Int Rev Cytol. 1995; 159:265–358.
Article4. de Crombrugghe B, Lefebvre V, Behringer RR, et al. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol. 2000; 19:389–394.
Article5. Hunziker EB, Kapfinger E, Geiss J. The structural architecture of adult mammalian articular cartilage evolves by a synchronized process of tissue resorption and neoformation during postnatal development. Osteoarthritis Cartilage. 2007; 15:403–413.
Article6. Hong E, Reddi AH. MicroRNAs in chondrogenesis, articular cartilage, and osteoarthritis: implications for tissue engineering. Tissue Eng Part B Rev. 2012; 18:445–453.
Article7. Kobayashi T, Lu J, Cobb BS, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A. 2008; 105:1949–1954.
Article8. Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005; 309:310–311.
Article9. Ason B, Darnell DK, Wittbrodt B, et al. Differences in vertebrate microRNA expression. Proc Natl Acad Sci U S A. 2006; 103:14385–14389.
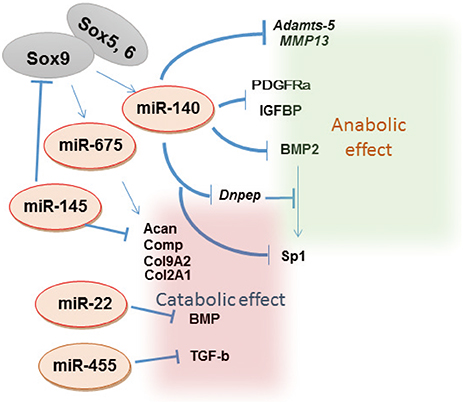
Article10. Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145). J Biol Chem. 2012; 287:916–924.
Article11. Nakamura Y, Inloes JB, Katagiri T, et al. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol. 2011; 31:3019–3028.
Article12. Miyaki S, Sato T, Inoue A, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010; 24:1173–1185.
Article13. Hong E, Reddi AH. Dedifferentiation and redifferentiation of articular chondrocytes from surface and middle zones: changes in microRNAs-221/-222, -140, and -143/145 expression. Tissue Eng Part A. 2013; 19:1015–1022.
Article14. Mow VC, Ratcliffe A, Poole AR. Cartilage and diarthrodial joints as paradigms for hierarchical materials and structures. Biomaterials. 1992; 13:67–97.
Article15. Månsson B, Carey D, Alini M, et al. Cartilage and bone metabolism in rheumatoid arthritis. Differences between rapid and slow progression of disease identified by serum markers of cartilage metabolism. J Clin Invest. 1995; 95:1071–1077.
Article16. Felson DT, Lawrence RC, Dieppe PA, et al. Osteoarthritis: new insights. Part 1: the disease and its risk factors. Ann Intern Med. 2000; 133:635–646.
Article17. Hashimoto M, Nakasa T, Hikata T, et al. Molecular network of cartilage homeostasis and osteoarthritis. Med Res Rev. 2008; 28:464–481.
Article18. Swingler TE, Wheeler G, Carmont V, et al. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 2012; 64:1909–1919.
Article19. Yang B, Kang X, Xing Y, et al. Effect of microRNA-145 on IL-1beta-induced cartilage degradation in human chondrocytes. FEBS Lett. 2014; 588:2344–2352.
Article20. Akhtar N, Rasheed Z, Ramamurthy S, et al. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 2010; 62:1361–1371.
Article21. Yamasaki K, Nakasa T, Miyaki S, et al. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009; 60:1035–1041.
Article22. Jones SW, Watkins G, Le Good N, et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthritis Cartilage. 2009; 17:464–472.
Article23. Lin EA, Kong L, Bai XH, et al. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009; 284:11326–11335.
Article24. Akhtar N, Haqqi TM. MicroRNA-199a* regulates the expression of cyclooxygenase-2 in human chondrocytes. Ann Rheum Dis. 2012; 71:1073–1080.
Article25. Matsukawa T, Sakai T, Yonezawa T, et al. MicroRNA-125b regulates the expression of aggrecanase-1 (ADAMTS-4) in human osteoarthritic chondrocytes. Arthritis Res Ther. 2013; 15:R28.
Article26. Mirzamohammadi F, Papaioannou G, Kobayashi T. MicroRNAs in cartilage development, homeostasis, and disease. Curr Osteoporos Rep. 2014; 12:410–419.
Article27. Papaioannou G, Mirzamohammadi F, Kobayashi T. MicroRNAs involved in bone formation. Cell Mol Life Sci. 2014; 71:4747–4761.
Article28. Barter MJ, Young DA. Epigenetic mechanisms and non-coding RNAs in osteoarthritis. Curr Rheumatol Rep. 2013; 15:353.
Article29. Gibson G, Asahara H. microRNAs and cartilage. J Orthop Res. 2013; 31:1333–1344.
Article30. Miyaki S, Asahara H. Macro view of microRNA function in osteoarthritis. Nat Rev Rheumatol. 2012; 8:543–552.
Article31. Clarkson MJ, Harley VR. Sex with two SOX on: SRY and SOX9 in testis development. Trends Endocrinol Metab. 2002; 13:106–111.
Article32. Tanaka K, Matsumoto Y, Nakatani F, et al. A zinc finger transcription factor, alphaA-crystallin binding protein 1, is a negative regulator of the chondrocyte-specific enhancer of the alpha1(II) collagen gene. Mol Cell Biol. 2006; 26:5202.
Article33. Lefebvre V, Huang W, Harley VR, et al. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997; 17:2336–2346.
Article34. Wagner T, Wirth J, Meyer J, et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994; 79:1111–1120.
Article35. Foster JW, Dominguez-Steglich MA, Guioli S, et al. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994; 372:525–530.
Article36. Zhao Q, Eberspaecher H, Lefebvre V, et al. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997; 209:377–386.
Article37. Bi W, Deng JM, Zhang Z, et al. Sox9 is required for cartilage formation. Nat Genet. 1999; 22:85–89.
Article38. Bi W, Huang W, Whitworth DJ, et al. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A. 2001; 98:6698–6703.
Article39. Murakami S, Lefebvre V, de Crombrugghe B. Potent inhibition of the master chondrogenic factor Sox9 gene by interleukin-1 and tumor necrosis factor-alpha. J Biol Chem. 2000; 275:3687–3692.
Article40. Nakamura Y, He X, Kato H, et al. Sox9 is upstream of microRNA-140 in cartilage. Appl Biochem Biotechnol. 2012; 166:64–71.
Article41. Yang J, Qin S, Yi C, et al. MiR-140 is co-expressed with Wwp2-C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett. 2011; 585:2992–2997.
Article42. Yamashita S, Miyaki S, Kato Y, et al. L-Sox5 and Sox6 proteins enhance chondrogenic miR-140 microRNA expression by strengthening dimeric Sox9 activity. J Biol Chem. 2012; 287:22206–22215.
Article43. Yang B, Guo H, Zhang Y, et al. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One. 2011; 6:e21679.
Article44. Licatalosi DD, Mele A, Fak JJ, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008; 456:464–469.
Article45. Takada S, Asahara H. Current strategies for microRNA research. Mod Rheumatol. 2012; 22:645–653.
Article46. Matsubara Y, Chiba T, Kashimada K, et al. Transcription activator-like effector nuclease-mediated transduction of exogenous gene into IL2RG locus. Sci Rep. 2014; 4:5043.
Article47. Takada S, Sato T, Ito Y, et al. Targeted gene deletion of miRNAs in mice by TALEN system. PLoS One. 2013; 8:e76004.
Article48. Inui M, Miyado M, Igarashi M, et al. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci Rep. 2014; 4:5396.
Article49. Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release. 2013; 172:962–974.
Article50. Ben-Shushan D, Markovsky E, Gibori H, et al. Overcoming obstacles in microRNA delivery towards improved cancer therapy. Drug Deliv Transl Res. 2014; 4:38–49.
Article