Isolation and characterization of human dental tissue-derived stem cells in the impacted wisdom teeth: comparison of dental follicle, dental pulp, and root apical papilla-derived cells
- Affiliations
-
- 1Department of Oral and Maxillofacial Surgery, School of Dentistry, Pusan National University, Yangsan, Korea. jorkim@pusan.ac.kr
- 2Department Oral and Maxillofacial Surgery, School of Medicine and Institute of Health Science, Gyeongsang National University, Jinju, Korea.
- 3College of Veterinary Medicine, Gyeongsang National University, Jinju, Korea.
- KMID: 2189946
- DOI: http://doi.org/10.5125/jkaoms.2010.36.3.186
Abstract
- INTRODUCTION
The first aim of this study was to isolate the dental tissue-derived stem cells from the dental follicle (DF), dental pulp (DP), and root apical papilla (RAP) of the extracted wisdom teeth. Second was to evaluate their characterization with the expressions of transcription factors and cell surface markers. Finally, their ability of the in vitro multi-lineage differentiations into osteogenic and adipogenic cells were compared, respectively.
MATERIALS AND METHODS
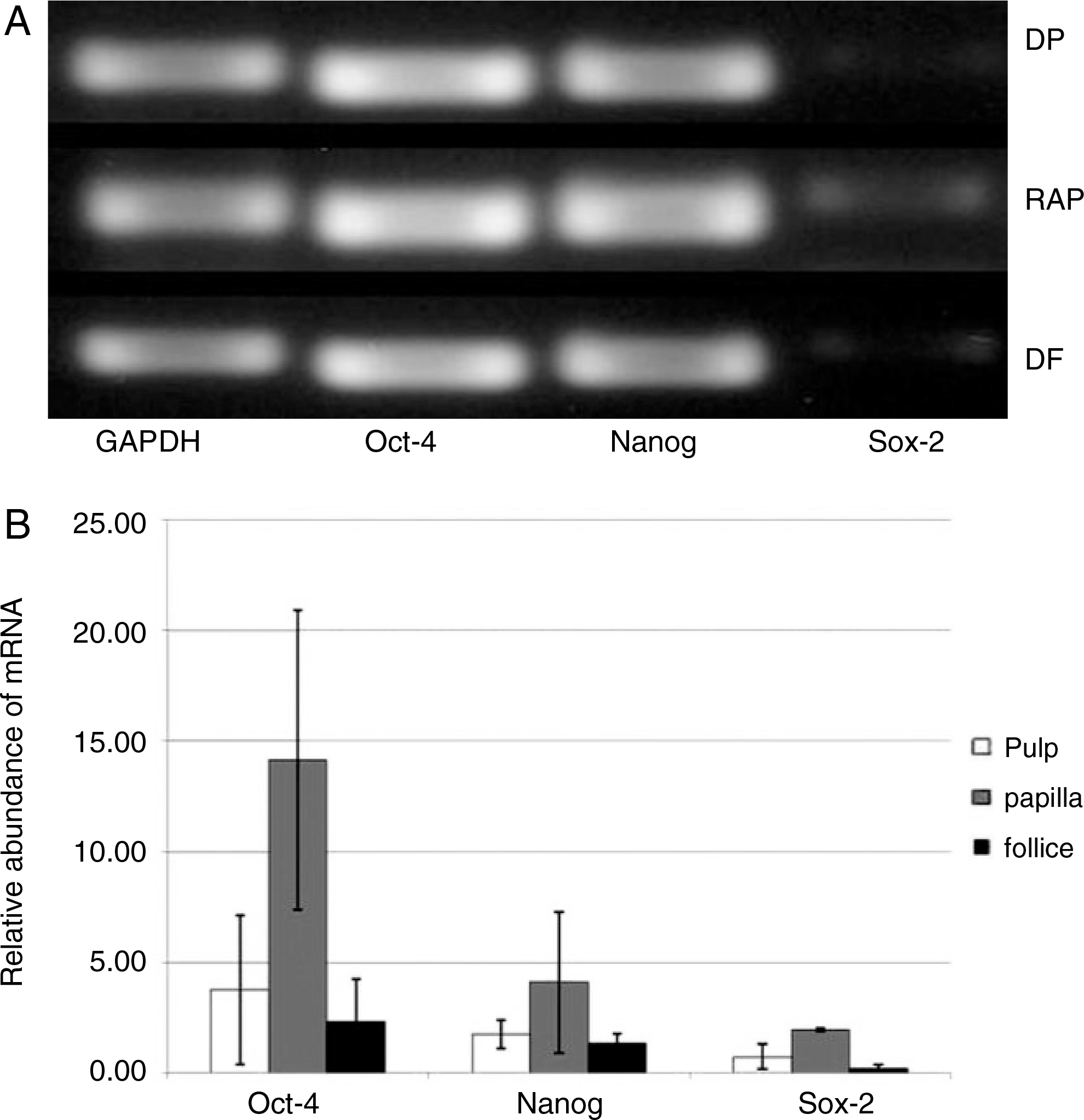
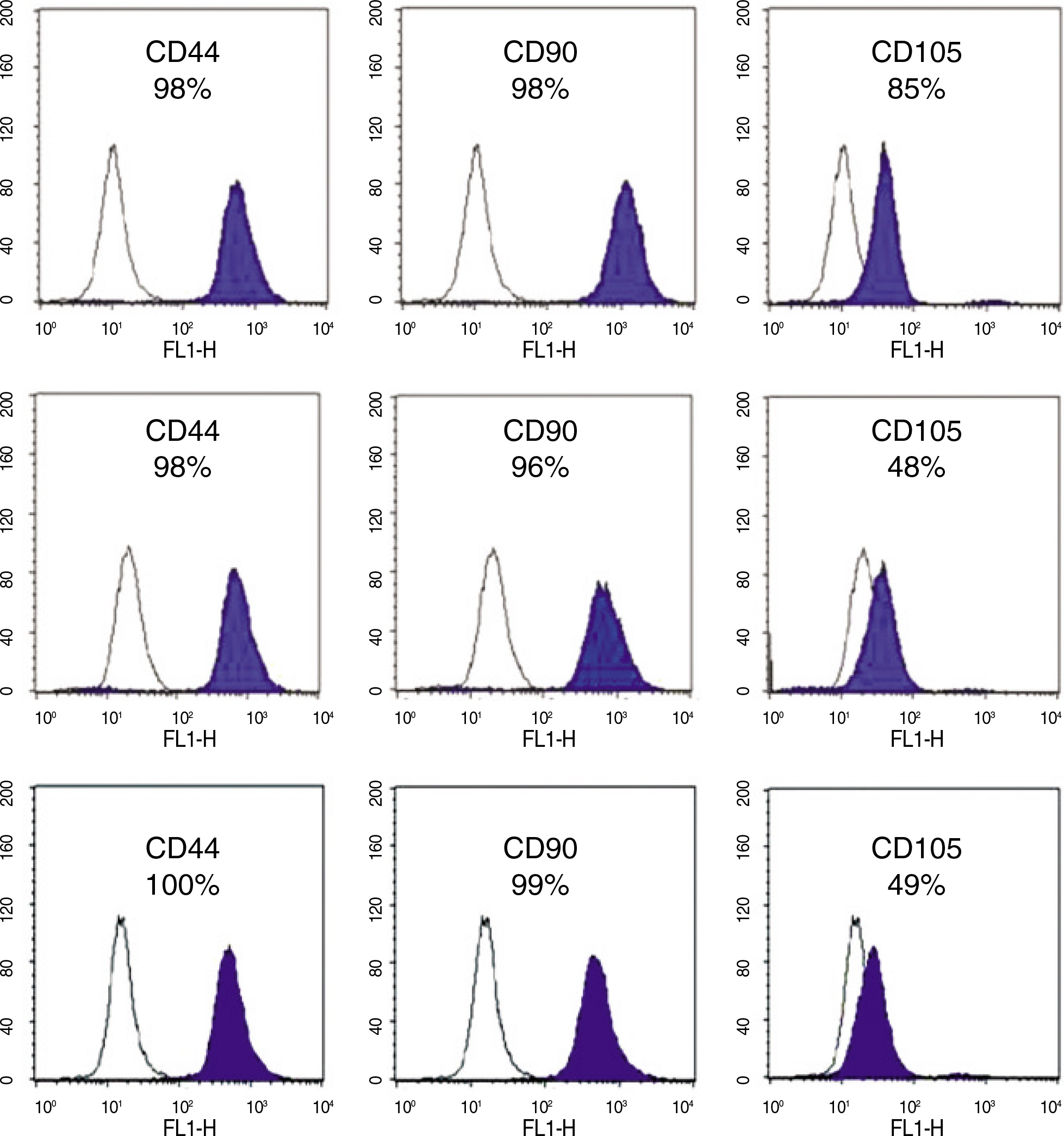
Dental tissues, including dental follicle, dental pulp, and root apical papilla, were separated in the extracted wisdom teeth. These three dental tissues were cultured in Dulbecco's modified Eagle's medium (DMEM) with supplements, respectively. After passage 3, the homogeneous shaped dental tissue-derived cells were analyzed the expression of transcription factors (Oct-4, Nanog and Sox-2) and cell surface markers (CD44, CD90 and CD105) with reverse transcription polymerase chain reaction (RT-PCR) and fluorescence-activated cell sorting (FACS) analysis. In order to evaluate in vitro multi-lineage differentiations, the culture media were changed to the osteogenic and adipogenic induction mediums when the dental tissue-derived cells reached to passage 3. The characteristics of these three dental tissue-derived cells were compared with immunohistochemistry.
RESULTS
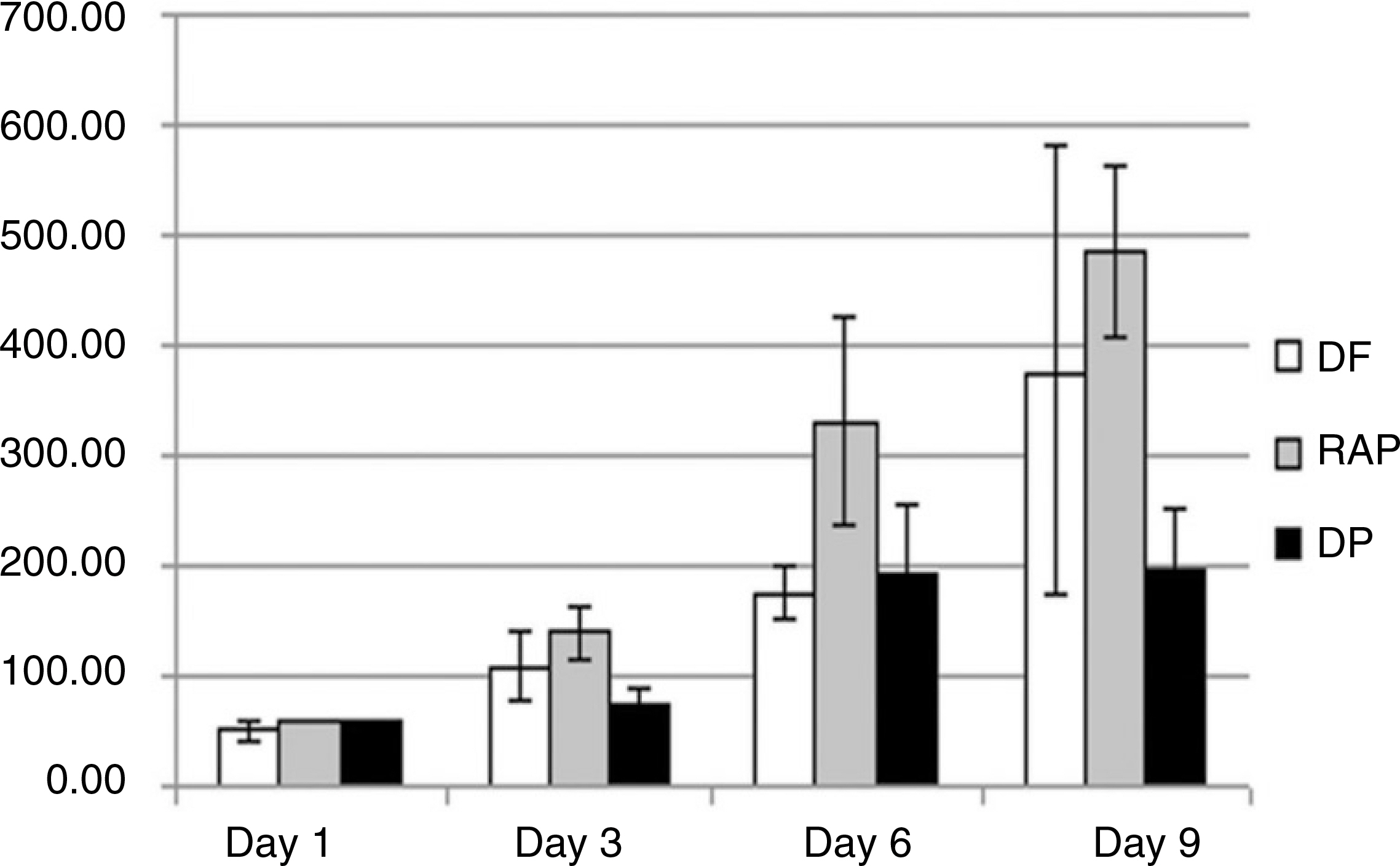
During primary culture, heterogenous and colony formatted dental tissue-derived cells were observed in the culture plates. After passage 2 or 3, homogenous spindle-like cells were observed in all culture plates. Transcription factors and mesenchymal stem cell markers were positively observed in all three types of dental tissue-derived cells. However, the quantity of expressed transcription factors was most large in RAP-derived cells. In all three types of dental tissue-derived cells, osteogenic and adipogenic differentiations were observed after treatment of specific induction media. In vitro adipogenic differentiation was similar among these three types of cells. In vitro osteogenic differentiation was most strongly and frequently observed in the RAP-derived cells, whereas rarely osteogenic differentiation was observed in the DP-derived cells.
CONCLUSION
These findings suggest that three types of human dental tissue-derived cells from extracted wisdom teeth were multipotent mesenchymal stem cells, have the properties of multi-lineage differentiations. Especially, stem cells from root apical papilla (SCAP) have much advantage in osteogenic differentiation, whereas dental follicle cells (DFCs) have a characteristic of easy adipogenic differentiation.
Keyword
MeSH Terms
Figure
Cited by 5 articles
-
Dental stem cells as a cell source for tissue engineering
Bong-Wook Park
J Korean Assoc Oral Maxillofac Surg. 2018;44(3):91-92. doi: 10.5125/jkaoms.2018.44.3.91.Regenerative medicine for the reconstruction of hard tissue defects in oral and maxillofacial surgery
Young-Kyun Kim
J Korean Assoc Oral Maxillofac Surg. 2012;38(2):69-70. doi: 10.5125/jkaoms.2012.38.2.69.Modulation of osteoblastic/odontoblastic differentiation of adult mesenchymal stem cells through gene introduction: a brief review
Ji-Youn Kim, Myung-Rae Kim, Sun-Jong Kim
J Korean Assoc Oral Maxillofac Surg. 2013;39(2):55-62. doi: 10.5125/jkaoms.2013.39.2.55.Dental tissues as adult stem cell source
Bong-Wook Park
J Korean Assoc Oral Maxillofac Surg. 2013;39(2):41-42. doi: 10.5125/jkaoms.2013.39.2.41.Cryopreservation of dental tissue and subsequent isolation of mesenchymal stem cells
Bong-Wook Park
J Korean Assoc Oral Maxillofac Surg. 2015;41(1):1-2. doi: 10.5125/jkaoms.2015.41.1.1.
Reference
-
References
1. Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981; 292:154–6.
Article2. Wagers AJ, Weissman IL. Plasticity of adult stem cells. Cell. 2004; 116:639–48.
Article3. Mao JJ, Giannobile WV, Helms JA, Hollister SJ, Krebsbach PH, Longaker MT, et al. Craniofacial tissue engineering by stem cells. J Dent Res. 2006; 85:966–79.
Article4. Almeida-Porada G, Porada C, Zanjani ED. Adult stem cell plasticity and methods of detection. Rev Clin Exp Hematol. 2001; 5:26–41.
Article5. Temple S. Stem cell plasticity-building the brain of our dreams. Nat Rev Neurosci. 2001; 2:513–20.6. Yamada Y, Boo JS, Ozawa R, Nagasaka T, Okazaki Y, Hata K, et al. Bone regeneration following injection of mesenchymal stem cells and fibrin glue with a biodegradable scaffold. J Craniomaxillofac Surg. 2003; 31:27–33.
Article7. Yamada Y, Ueda M, Naiki T, Nagasaka T. Tissue-engineered injectable bone regeneration for osseointegrated dental implants. Clin Oral Impl Res. 2004; 15:589–97.
Article8. Fuerst G, Tangl S, Gruber R, Gahleitner A, Sanroman F, Watzek G. Bone formation following sinus grafting with autogenous bone-derived cells and bovine bone mineral in minipigs: preliminary findings. Clin Oral Impl Res. 2004; 15:733–40.
Article9. Schimming R, Schmelzeisen R. Tissue-engineered bone for maxillary sinus augmentation. J Oral Maxillofac Surg. 2004; 62:724–9.
Article10. Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA. 2000; 97:13625–30.11. Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A, et al. Stem cell properties of human dental pulp stem cells. J Dent Res. 2002; 81:531–5.
Article12. Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, et al. Stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA. 2003; 100:5807–12.13. Seo BM, Sonoyama W, Yamaza T, Coppe C, Kikuiri T, Akiyama K, et al. SHED repair critical-size calvarial defects in mice. Oral Dis. 2008; 14:428–34.
Article14. Huang AH, Chen YK, Lin LM, Shieh TY, Chan AW. Isolation and characterization of dental pulp stem cells from a supernumerary tooth. J Oral Pathol Med. 2008; 37:571–4.
Article15. Morsczeck C, Go ¨tz W, Schierholz J, Zeilhofer F, Ku ¨hn U, Mo ¨hl C, et al. Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 2005; 24:155–65.
Article16. Morsczeck C, Moehl C, Go ¨tz W, Heredia A, Scha ¨ffer TE, Eckstein N, et al. In vitro differentiation of human dental follicle cells with dexamethasone and insulin. Cell Biol Int. 2005; 29:567–75.
Article17. Reynolds AJ, Jahoda CA. Cultured human and rat tooth papilla cells induce hair follicle regeneration and fiber growth. Differentiation. 2004; 72:566–75.
Article18. Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004; 364:149–55.
Article19. Sonoyama W, Liu Y, Fang D, Yamaza T, Seo BM, Zhang C, et al. Mesenchymal stem cell-mediated functional tooth regeneration in swine. PLoS ONE. 2006; 1:e79.
Article20. Morsczeck C, Vo ¨llner F, Saugspier M, Brandl C, Reichert TE, Driemel O, et al. Comparison of human dental follicle cells (DFCs) and stem cells from human exfoliated deciduous teeth (SHED) after neural differentiation in vitro. Clin Oral Investig [in press 2009 Jul 10].21. Kang EJ, Byun JH, Choi YJ, Maeng GH, Lee SL, Kang DH, et al. In vitro and in vivo osteogenesis of porcine skin-derived mesenchymal stem cell-like cells with a demineralized bone and fibrin scaffold. Tissue Eng Part A. 2010; 16:815–27.22. Scho ¨ler HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990; 344:435–9.
Article23. Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003; 17:126–40.
Article24. Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie .
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparison of Gene Expression from Supernumerary Dental Pulp and Periodontal Ligament Stem Cells
- Characterization of Human Dental Pulp Cells from Supernumerary Teeth by Using Flow Cytometry Analysis
- Modulation of osteoblastic/odontoblastic differentiation of adult mesenchymal stem cells through gene introduction: a brief review
- A study on differentiation potency of adult stem cells from pulp, periodontal ligament, and dental follicle to osteoblast
- Induced Pluripotent Stem (iPS) Cells in Dentistry: A Review