J Adv Prosthodont.
2015 Dec;7(6):468-474. 10.4047/jap.2015.7.6.468.
Partial denture metal framework may harbor potentially pathogenic bacteria
- Affiliations
-
- 1Department of Conservative Dentistry, Federal University of Rio Grande do Sul, School of Dentistry, Porto Alegre, Rio Grande do Sul, Brazil. cristiane.mengatto@ufrgs.br
- 2Department of Preventive and Community Dentistry, University of Iowa, College of Dentistry, Iowa City, Iowa, USA.
- 3Department of Science and Technology, State University of Santa Cruz, School of Computer Science, Ilheus, Bahia, Brazil.
- 4Department of Dental Materials and Prosthodontics, State University Julio de Mesquita Filho, School of Dentistry, Sao Jose dos Campos, Sao Paulo, Brazil.
- 5Department of Prosthodontics and Periodontology, State University of Campinas, Piracicaba Dental School, Piracicaba, Sao Paulo, Brazil.
- KMID: 2176622
- DOI: http://doi.org/10.4047/jap.2015.7.6.468
Abstract
- PURPOSE
The aim of this study was to characterize and compare bacterial diversity on the removable partial denture (RPD) framework over time.
MATERIALS AND METHODS
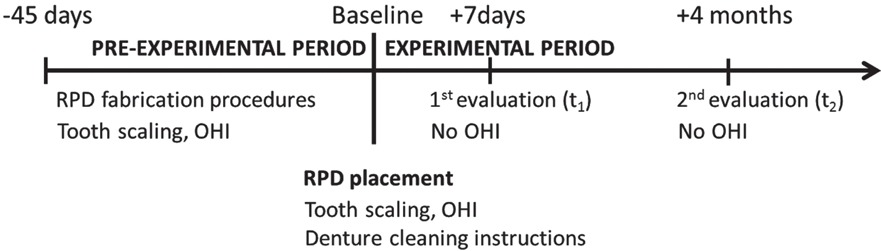
This descriptive pilot study included five women who were rehabilitated with free-end mandibular RPD. The biofilm on T-bar clasps were collected 1 week (t1) and 4 months (t2) after the RPD was inserted (t0). Bacterial 16S rDNA was extracted and PCR amplified. Amplicons were cloned; clones were submitted to cycle sequencing, and sequences were compared with GenBank (98% similarity).
RESULTS
A total of 180 sequences with more than 499 bp were obtained. Two phylogenetic trees with 84 (t1) and 96 (t2) clones represented the bacteria biofilm at the RPD. About 93% of the obtained phylotypes fell into 25 known species for t1 and 17 for t2, which were grouped in 5 phyla: Firmicutes (t1=82%; t2=60%), Actinobacteria (t1=5%; t2=10%), Bacteroidetes (t1=2%; t2=6%), Proteobacteria (t1=10%; t2=15%) and Fusobacteria (t1=1%; t2=8%). The libraries also include 3 novel phylotypes for t1 and 11 for t2. Library t2 differs from t1 (P=.004); t1 is a subset of the t2 (P=.052). Periodontal pathogens, such as F. nucleatum, were more prevalent in t2.
CONCLUSION
The biofilm composition of the RPD metal clasps changed along time after RPD wearing. The RPD framework may act as a reservoir for potentially pathogenic bacteria and the RPD wearers may benefit from regular follow-up visits and strategies on prosthesis-related oral health instructions.
MeSH Terms
Figure
Reference
-
1. Bergman B, Hugoson A, Olsson CO. A 25 year longitudinal study of patients treated with removable partial dentures. J Oral Rehabil. 1995; 22:595–599.2. Mullally BH, Linden GJ. Periodontal status of regular dental attenders with and without removable partial dentures. Eur J Prosthodont Restor Dent. 1994; 2:161–163.3. Schwalm CA, Smith DE, Erickson JD. A clinical study of patients 1 to 2 years after placement of removable partial dentures. J Prosthet Dent. 1977; 38:380–391.4. Zlatarić DK, Celebić A, Valentić-Peruzović M. The effect of removable partial dentures on periodontal health of abutment and non-abutment teeth. J Periodontol. 2002; 73:137–144.5. do Amaral BA, Barreto AO, Gomes Seabra E, Roncalli AG, da Fonte Porto Carreiro A, de Almeida EO. A clinical follow-up study of the periodontal conditions of RPD abutment and non-abutment teeth. J Oral Rehabil. 2010; 37:545–552.6. Vanzeveren C, D'Hoore W, Bercy P. Influence of removable partial denture on periodontal indices and microbiological status. J Oral Rehabil. 2002; 29:232–239.7. Zhu X, Wang S, Gu Y, Li X, Yan H, Yan H, Miyoshi S, Shi L. Possible variation of the human oral bacterial community after wearing removable partial dentures by DGGE. World J Microbiol Biotechnol. 2012; 28:2229–2236.8. Campos MS, Marchini L, Bernardes LA, Paulino LC, Nobrega FG. Biofilm microbial communities of denture stomatitis. Oral Microbiol Immunol. 2008; 23:419–424.9. Danser MM, van Winkelhoff AJ, de Graaff J, van der Velden U. Putative periodontal pathogens colonizing oral mucous membranes in denture-wearing subjects with a past history of periodontitis. J Clin Periodontol. 1995; 22:854–859.10. Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ Jr, Kolenbrander PE. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006; 72:2837–2848.11. Fernandes CB, Aquino DR, Franco GC, Cortelli SC, Costa FO, Cortelli JR. Do elderly edentulous patients with a history of periodontitis harbor periodontal pathogens? Tex Dent J. 2012; 129:751–761.12. Hutter G, Schlagenhauf U, Valenza G, Horn M, Burgemeister S, Claus H, Vogel U. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology. 2003; 149:67–75.13. Ledder RG, Gilbert P, Huws SA, Aarons L, Ashley MP, Hull PS, McBain AJ. Molecular analysis of the subgingival microbiota in health and disease. Appl Environ Microbiol. 2007; 73:516–523.14. Mantzourani M, Gilbert SC, Fenlon M, Beighton D. Non-oral bifidobacteria and the aciduric microbiota of the denture plaque biofilm. Mol Oral Microbiol. 2010; 25:190–199.15. Marchini L, Campos MS, Silva AM, Paulino LC, Nobrega FG. Bacterial diversity in aphthous ulcers. Oral Microbiol Immunol. 2007; 22:225–231.16. Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001; 183:3770–3783.17. Sachdeo A, Haffajee AD, Socransky SS. Biofilms in the edentulous oral cavity. J Prosthodont. 2008; 17:348–356.18. Wade WG, Spratt DA, Dymock D, Weightman AJ. Molecular detection of novel anaerobic species in dentoalveolar abscesses. Clin Infect Dis. 1997; 25:S235–S236.19. Mihalow DM, Tinanoff N. The influence of removable partial dentures on the level of Streptococcus mutans in saliva. J Prosthet Dent. 1988; 59:49–51.20. Rocha EP, Francisco SB, Del Bel Cury AA, Cury JA. Longitudinal study of the influence of removable partial denture and chemical control on the levels of Streptococcus mutans in saliva. J Oral Rehabil. 2003; 30:131–138.21. Yasui M, Ryu M, Sakurai K, Ishihara K. Colonisation of the oral cavity by periodontopathic bacteria in complete denture wearers. Gerodontology. 2012; 29:e494–e502.22. Grivet M, Morrier JJ, Benay G, Barsotti O. Effect of hydrophobicity on in vitro streptococcal adhesion to dental alloys. J Mater Sci Mater Med. 2000; 11:637–642.23. Mine K, Fueki K, Igarashi Y. Microbiological risk for periodontitis of abutment teeth in patients with removable partial dentures. J Oral Rehabil. 2009; 36:696–702.24. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005; 43:5721–5732.25. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010; 192:5002–5017.26. Urushibara Y, Ohshima T, Sato M, Hayashi Y, Hayakawa T, Maeda N, Ohkubo C. An analysis of the biofilms adhered to framework alloys using in vitro denture plaque models. Dent Mater J. 2014; 33:402–414.27. Carr AB, McGivney GP, Brown DT, McCracken WL. McCracken's Removable Partial Prosthodontics. 12 ed. St. Louis: Mosby;2010.28. Chao A. Non-parametric estimation of the number of classes in a population. Scand J Stat. 1984; 11:265–270.29. Chao A, Lee SM. Estimating the number of classes via sample coverage. J Am Stat Assoc. 1992; 87:210–217.30. Marsh PD, Percival RS, Challacombe SJ. The influence of denture-wearing and age on the oral microflora. J Dent Res. 1992; 71:1374–1381.31. Nyvad B, Kilian M. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand J Dent Res. 1987; 95:369–380.32. Taylor R, Maryan C, Verran J. Retention of oral microorganisms on cobalt-chromium alloy and dental acrylic resin with different surface finishes. J Prosthet Dent. 1998; 80:592–597.33. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998; 25:134–144.34. Siqueira JF Jr, Rôças IN. Catonella morbi and Granulicatella adiacens: new species in endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006; 102:259–264.35. Gonçalves LF, Fermiano D, Feres M, Figueiredo LC, Teles FR, Mayer MP, Faveri M. Levels of Selenomonas species in generalized aggressive periodontitis. J Periodontal Res. 2012; 47:711–718.36. Cone LA, Leung MM, Hirschberg J. Actinomyces odontolyticus bacteremia. Emerg Infect Dis. 2003; 9:1629–1632.37. Haffajee AD, Teles RP, Patel MR, Song X, Veiga N, Socransky SS. Factors affecting human supragingival biofilm composition. I. Plaque mass. J Periodontal Res. 2009; 44:511–519.38. Rüdiger SG, Carlén A, Meurman JH, Kari K, Olsson J. Dental biofilms at healthy and inflamed gingival margins. J Clin Periodontol. 2002; 29:524–530.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Cr-Co removable partial denture treatment fabricated by selective laser melting: a case report
- Comparison of internal adaptation of removable partial denture metal frameworks made by lost wax technique and printing technique of pattern using CAD
- A case of removable dentures using digital method
- Complete denture making in a patient of partial glossectomy using polished surface impression taking and direct metal laser sintering method: A case report
- RPD framework fabrication using computer-aided design (CAD) and rapid prototyping