J Breast Cancer.
2015 Dec;18(4):329-338. 10.4048/jbc.2015.18.4.329.
Synergistic Effect of Trabectedin and Olaparib Combination Regimen in Breast Cancer Cell Lines
- Affiliations
-
- 1Cell Biology and Pharmacogenomics Department, PharmaMar S.A., Madrid, Spain. cgalmarini@pharmamar.com
- KMID: 2176281
- DOI: http://doi.org/10.4048/jbc.2015.18.4.329
Abstract
- PURPOSE
Trabectedin induces synthetic lethality in tumor cells carrying defects in homologous recombinant DNA repair. We evaluated the effect of concomitant inhibition of nucleotide-excision repair and poly (ADP-ribose) polymerase (PARP) activity with trabectedin and PARP inhibitors, respectively, and whether the synthetic lethality effect had the potential for a synergistic effect in breast cancer cell lines. Additionally, we investigated if this approach remained effective in BRCA1-positive breast tumor cells.
METHODS
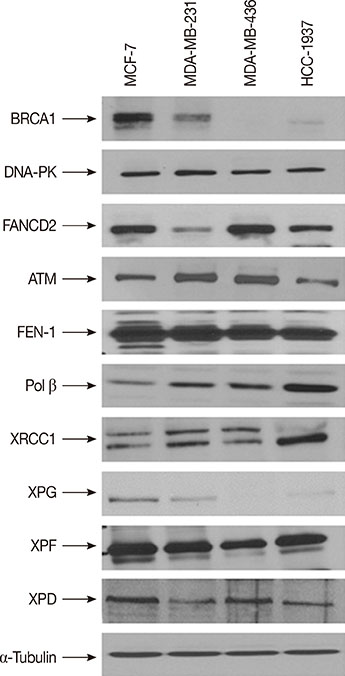
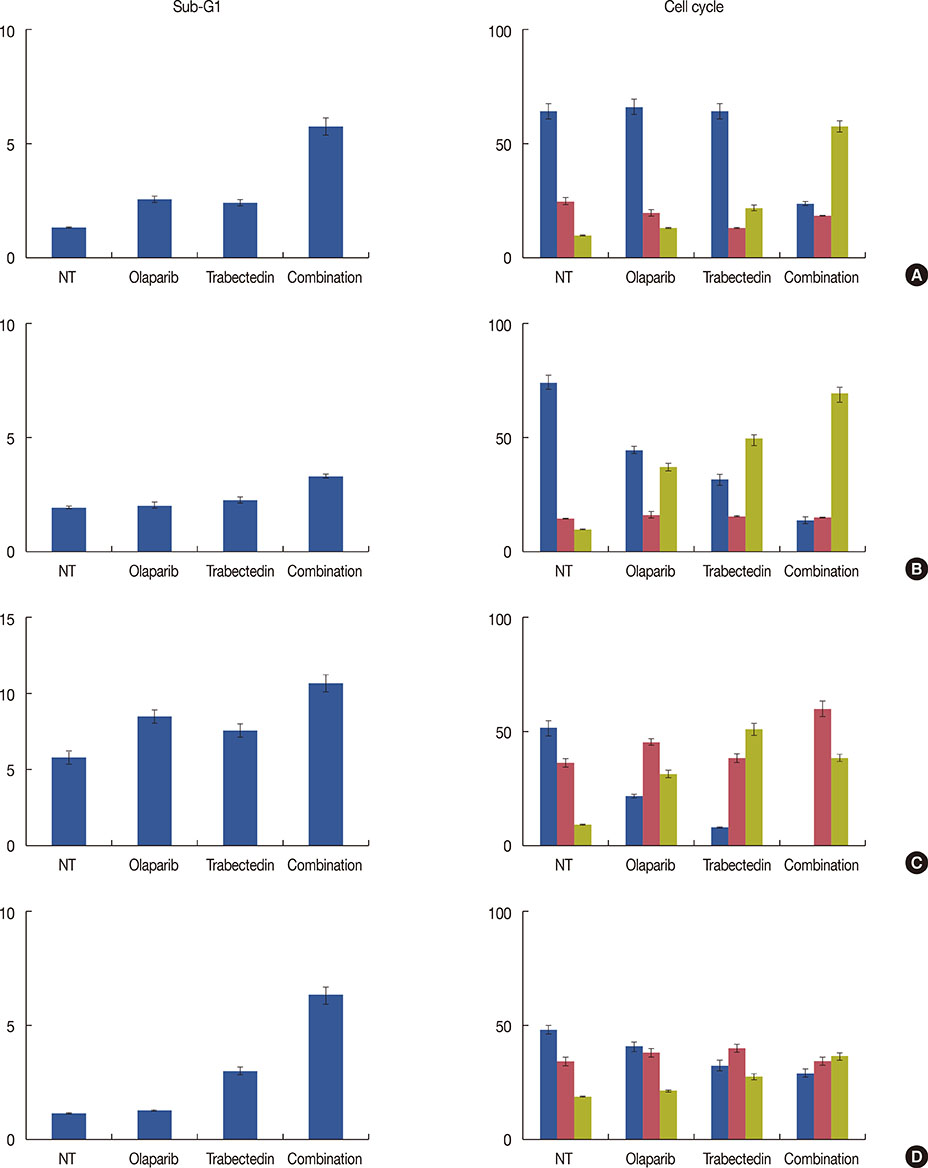
We have evaluated the in vitro synergistic effect of combinations of trabectedin and three different PARP inhibitors (veliparib, olaparib, and iniparib) in four breast cancer cell lines, each presenting a different BRCA1 genetic background. Antiproliferative activity, DNA damage, cell cycle perturbations and poly(ADP-ribosyl)ation were assessed by MTT assay, comet assay, flow cytometry and western blot, respectively.
RESULTS
The combination of trabectedin and olaparib was synergistic in all the breast cancer cell lines tested. Our data indicated that the synergy persisted regardless of the BRCA1 status of the tumor cells. Combination treatment was associated with a strong accumulation of double-stranded DNA breaks, G2/M arrest, and apoptotic cell death. Synergistic effects were not observed when trabectedin was combined with veliparib or iniparib.
CONCLUSION
Collectively, our results indicate that the combination of trabectedin and olaparib induces an artificial synthetic lethality effect that can be used to kill breast cancer cells, independent of BRCA1 status.
MeSH Terms
Figure
Reference
-
1. D'Incalci M, Galmarini CM. A review of trabectedin (ET-743): a unique mechanism of action. Mol Cancer Ther. 2010; 9:2157–2163.2. Demetri GD, Chawla SP, von Mehren M, Ritch P, Baker LH, Blay JY, et al. Efficacy and safety of trabectedin in patients with advanced or metastatic liposarcoma or leiomyosarcoma after failure of prior anthracyclines and ifosfamide: results of a randomized phase II study of two different schedules. J Clin Oncol. 2009; 27:4188–4196.
Article3. Monk BJ, Herzog TJ, Kaye SB, Krasner CN, Vermorken JB, Muggia FM, et al. Trabectedin plus pegylated liposomal doxorubicin in recurrent ovarian cancer. J Clin Oncol. 2010; 28:3107–3114.
Article4. Di Giandomenico S, Frapolli R, Bello E, Uboldi S, Licandro SA, Marchini S, et al. Mode of action of trabectedin in myxoid liposarcomas. Oncogene. 2014; 33:5201–5210.
Article5. Galmarini CM, D'Incalci M, Allavena P. Trabectedin and plitidepsin: drugs from the sea that strike the tumor microenvironment. Mar Drugs. 2014; 12:719–733.
Article6. Herrero AB, Martín-Castellanos C, Marco E, Gago F, Moreno S. Crosstalk between nucleotide excision and homologous recombination DNA repair pathways in the mechanism of action of antitumor trabectedin. Cancer Res. 2006; 66:8155–8162.
Article7. Feuerhahn S, Giraudon C, Martínez-Díez M, Bueren-Calabuig JA, Galmarini CM, Gago F, et al. XPF-dependent DNA breaks and RNA polymerase II arrest induced by antitumor DNA interstrand crosslinking-mimetic alkaloids. Chem Biol. 2011; 18:988–999.
Article8. Takebayashi Y, Pourquier P, Zimonjic DB, Nakayama K, Emmert S, Ueda T, et al. Antiproliferative activity of ecteinascidin 743 is dependent upon transcription-coupled nucleotide-excision repair. Nat Med. 2001; 7:961–966.
Article9. Soares DG, Poletto NP, Bonatto D, Salvador M, Schwartsmann G, Henriques JA. Low cytotoxicity of ecteinascidin 743 in yeast lacking the major endonucleolytic enzymes of base and nucleotide excision repair pathways. Biochem Pharmacol. 2005; 70:59–69.
Article10. Romano M, Frapolli R, Zangarini M, Bello E, Porcu L, Galmarini CM, et al. Comparison of in vitro and in vivo biological effects of trabectedin, lurbinectedin (PM01183) and Zalypsis(R) (PM00104). Int J Cancer. 2013; 133:2024–2033.
Article11. Soares DG, Escargueil AE, Poindessous V, Sarasin A, de Gramont A, Bonatto D, et al. Replication and homologous recombination repair regulate DNA double-strand break formation by the antitumor alkylator ecteinascidin 743. Proc Natl Acad Sci U S A. 2007; 104:13062–13067.
Article12. Nijman SM, Friend SH. Cancer: potential of the synthetic lethality principle. Science. 2013; 342:809–811.13. Garber K. Synthetic lethality: killing cancer with cancer. J Natl Cancer Inst. 2002; 94:1666–1668.
Article14. Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat Med. 2013; 19:1381–1388.
Article15. Helleday T. Putting poly (ADP-ribose) polymerase and other DNA repair inhibitors into clinical practice. Curr Opin Oncol. 2013; 25:609–614.
Article16. Elstrodt F, Hollestelle A, Nagel JH, Gorin M, Wasielewski M, van den Ouweland A, et al. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006; 66:41–45.
Article17. Peralta-Leal A, Rodríguez-Vargas JM, Aguilar-Quesada R, Rodríguez MI, Linares JL, de Almodóvar MR, et al. PARP inhibitors: new partners in the therapy of cancer and inflammatory diseases. Free Radic Biol Med. 2009; 47:13–26.
Article18. Horton TM, Jenkins G, Pati D, Zhang L, Dolan ME, Ribes-Zamora A, et al. Poly(ADP-ribose) polymerase inhibitor ABT-888 potentiates the cytotoxic activity of temozolomide in leukemia cells: influence of mismatch repair status and O6-methylguanine-DNA methyltransferase activity. Mol Cancer Ther. 2009; 8:2232–2242.
Article19. McPherson LA, Shen Y, Ford JM. Poly (ADP-ribose) polymerase inhibitor LT-626: sensitivity correlates with MRE11 mutations and synergizes with platinums and irinotecan in colorectal cancer cells. Cancer Lett. 2014; 343:217–223.
Article20. Vilar E, Bartnik CM, Stenzel SL, Raskin L, Ahn J, Moreno V, et al. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res. 2011; 71:2632–2642.
Article21. Anderson VE, Walton MI, Eve PD, Boxall KJ, Antoni L, Caldwell JJ, et al. CCT241533 is a potent and selective inhibitor of CHK2 that potentiates the cytotoxicity of PARP inhibitors. Cancer Res. 2011; 71:463–472.
Article22. Lord CJ, McDonald S, Swift S, Turner NC, Ashworth A. A highthroughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair (Amst). 2008; 7:2010–2019.
Article23. Cheng H, Zhang Z, Borczuk A, Powell CA, Balajee AS, Lieberman HB, et al. PARP inhibition selectively increases sensitivity to cisplatin in ERCC1-low non-small cell lung cancer cells. Carcinogenesis. 2013; 34:739–749.
Article24. Chuang HC, Kapuriya N, Kulp SK, Chen CS, Shapiro CL. Differential anti-proliferative activities of poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative breast cancer cells. Breast Cancer Res Treat. 2012; 134:649–659.
Article25. Hastak K, Alli E, Ford JM. Synergistic chemosensitivity of triple-negative breast cancer cell lines to poly(ADP-Ribose) polymerase inhibition, gemcitabine, and cisplatin. Cancer Res. 2010; 70:7970–7980.
Article26. Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012; 72:5588–5599.
Article27. D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl) ation reactions in the regulation of nuclear functions. Biochem J. 1999; 342(Pt 2):249–268.28. Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011; 41:210–220.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Cellular Effect of Curcumin and Citral Combination on Breast Cancer Cells: Induction of Apoptosis and Cell Cycle Arrest
- Synergistic Cytotoxic Effects of Zole-dronic Acid and Radiation in the MCF-7 Human Breast Cancer Cell Line
- Synergistic Effect of Flavonoids from Artocarpus heterophyllus Heartwoods on Anticancer Activity of Cisplatin Against H460 and MCF-7 Cell Lines
- Inhibition of WEE1 Potentiates Sensitivity to PARP Inhibitor in Biliary Tract Cancer
- In Vitro Interaction of Taxol with Other Antitumor Drugs in the Established Choriocarcinoma Cell Lines