Chonnam Med J.
2009 Apr;45(1):19-26. 10.4068/cmj.2009.45.1.19.
Multiplex PCR Assay for Identification of Mycobacterial Species Isolated from Liquid Cultures
- Affiliations
-
- 1Department of Laboratory Medicine, Chonnam National University Medical School, Gwangju, Korea. spsuh@chonnam.ac.kr
- 2Department of Internal Medicine, Chonnam National University Medical School, Gwangju, Korea.
- KMID: 2172279
- DOI: http://doi.org/10.4068/cmj.2009.45.1.19
Abstract
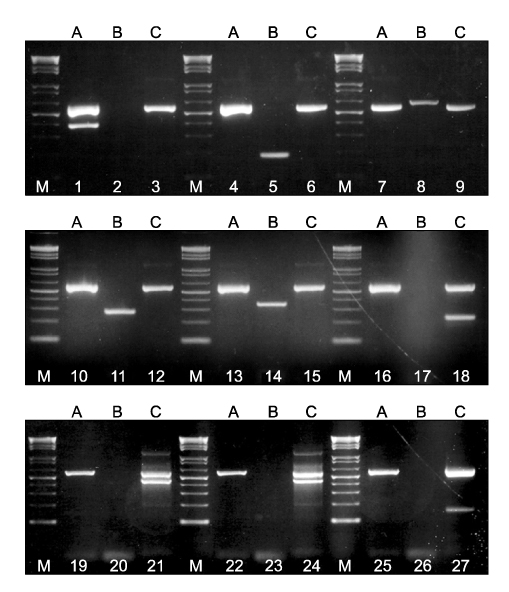
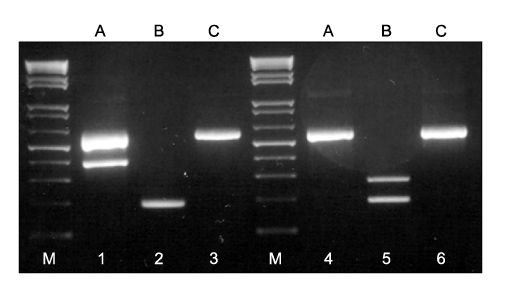
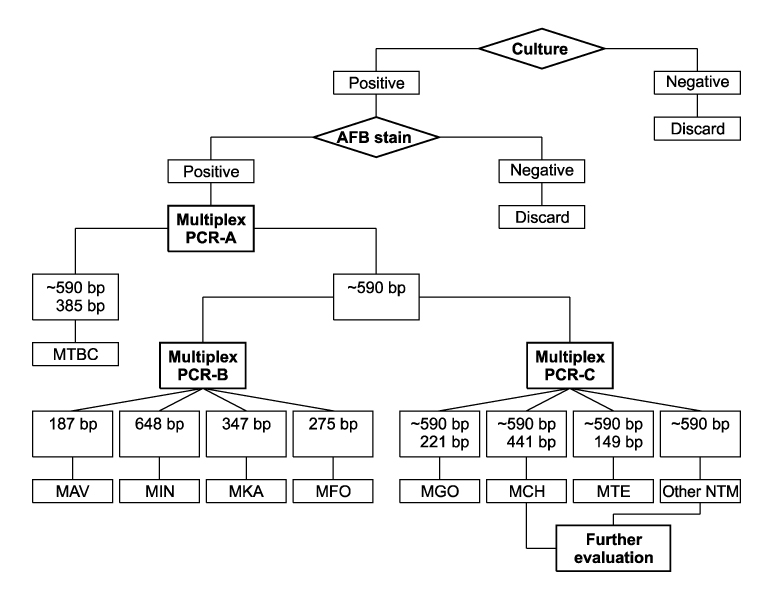
- We developed a 2-step multiplex polymerase chain reaction (PCR) assay to rapidly identify the most common mycobacterial species isolated from positive liquid cultures as follows. In the first step, a multiplex PCR-A assay was used to differentiate Mycobacterium tuberculosis complex from nontuberculous mycobacterial (NTM) species. In the second step, 2 parallel multiplex PCR-B/C assays were used to identify M. avium, M. intracellulare, M. kansasii, M. fortuitum, M. abscessus/chelonae, M. gordonae, and M. terrae. The multiplex PCR assays were tested on a total of 147 liquid cultures, including 15 reference strains and 4 mixed cultures. The results were compared with those obtained by PCR-restriction enzyme analysis of the hsp65 gene (PRA-hsp65) and species-specific PCR. For 129 of 143 cultures yielding a single isolate, the results of the multiplex PCR matched those of species-specific PCR and PRA-hsp65 except for 8 M. intracellulare isolates and 6 other NTM species not represented by the multiplex PCR in this study. Moreover, the multiplex PCR detected the coexistence of 2 mycobacterial species in 4 mixed cultures, whereas PRA-hsp65 misidentified the mixed cultures as a single species. The data demonstrated that multiplex PCR assays may be easy and useful for the rapid identification of most common mycobacterial species in liquid cultures, particularly in mixed cultures.
MeSH Terms
Figure
Reference
-
1. Hong YP, Kim SJ, Lew WJ, Lee EK, Han YC. The seventh nationwide tuberculosis prevalence survey in Korea, 1995. Int J Tuberc Lung Dis. 1998. 2:27–36.2. Hanna BA, Ebrahimzadeh A, Elliott LB, Morgan MA, Novak SM, Rusch-Gerdes S, et al. Multicenter evaluation of the BACTEC MGIT 960 system for recovery of mycobacteria. J Clin Microbiol. 1999. 37:748–752.
Article3. Butler WR, Guthertz LS. Mycolic acid analysis by high-performance liquid chromatography for identification of Mycobacterium species. Clin Microbiol Rev. 2001. 14:704–726.
Article4. Walton DT, Valesco M. Identification of Mycobacterium gordonae from culture by the Gen-Probe Rapid Diagnostic System: evaluation of 218 isolates and potential sources of false-negative results. J Clin Microbiol. 1991. 29:1850–1854.
Article5. Tortoli E, Simonetti MT, Lavinia F. Evaluation of reformulated chemiluminescent DNA probe (AccuProbe) for culture identification of Mycobacterium kansasii. J Clin Microbiol. 1996. 34:2838–2840.
Article6. Devallois A, Picardeau M, Paramasivan CN, Vincent V, Rastogi N. Molecular characterization of Mycobacterium avium complex isolates giving discordant results in AccuProbe tests by PCR-restriction enzyme analysis, 16S rRNA gene sequencing, and DT1-DT6 PCR. J Clin Microbiol. 1997. 35:2767–2772.
Article7. Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, et al. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993. 31:2882–2889.
Article8. Daley P, Petrich A, May K, Luinstra K, Rutherford C, Chedore P, et al. Comparison of in-house and commercial 16S rRNA sequencing with high-performance liquid chromatography and genotype AS and CM for identification of nontuberculous mycobacteria. Diagn Microbiol Infect Dis. 2008. 61:284–293.
Article9. Wilton S, Cousins D. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. PCR Methods Appl. 1992. 1:269–273.
Article10. Cousins D, Francis B, Dawson D. Multiplex PCR provides a low-cost alternative to DNA probe methods for rapid identification of Mycobacterium avium and Mycobacterium intracellulare. J Clin Microbiol. 1996. 34:2331–2333.
Article11. Thierry D, Vincent V, Clément F, Guesdon JL. Isolation of specific DNA fragments of Mycobacterium avium and their possible use in diagnosis. J Clin Microbiol. 1993. 31:1048–1054.
Article12. Yang M, Ross BC, Dwyer B. Isolation of a DNA probe for identification of Mycobacterium kansasii, including the genetic subgroup. J Clin Microbiol. 1993. 31:2769–2772.
Article13. Picardeau M, Prod'Hom G, Raskine L, LePennec MP, Vincent V. Genotypic characterization of five subspecies of Mycobacterium kansasii. J Clin Microbiol. 1997. 35:25–32.
Article14. Zolg JW, Philippi-Schulz S. The superoxide dismutase gene, a target for detection and identification of mycobacteria by PCR. J Clin Microbiol. 1994. 32:2801–2812.
Article15. Klemen H, Bogiatzis A, Ghalibafian M, Popper HH. Multiplex polymerase chain reaction for rapid detection of atypical mycobacteria and Mycobacterium tuberculosis complex. Diagn Mol Pathol. 1998. 7:310–316.
Article16. Picardeau M, Varnerot A, Rauzier J, Gicquel B, Vincent V. Mycobacterium xenopi IS1395, a novel insertion sequence expanding the IS256 family. Microbiology. 1996. 142:2453–2461.
Article17. Ryang DW, Ryang DH, Shin MG, Shin JH, Kee SJ, Suh SP. Alternative use of polymerase chain reaction instead of rho-nitro-alpha-acetylamino-beta-hydroxypropiophenone test for the early detection of Mycobacterium tuberculosis in BACTEC 12B cultures. APMIS. 1996. 104:444–450.
Article18. Edwards MC, Gibs RA. Dieffenbach CW, Dveksler GS, editors. Multiplex PCR. PCR PRIMER: a laboratory manual. 1995. 1ST ed. New York: Cold Spring Harbor Laboratory Press;157–171.19. Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993. 31:175–178.
Article20. Chimara E, Ferrazoli L, Ueky SY, Martins MC, Durham AM, Arbeit RD, et al. Reliable identification of mycobacterial species by PCR-restriction enzyme analysis (PRA)-hsp65 in a reference laboratory and elaboration of a sequence-based extended algorithm of PRA-hsp65 patterns. BMC Microbiol. 2008. 8:48.21. Ben Salah I, Adékambi T, Raoult D, Drancourt M. rpoB sequence-based identification of Mycobacterium avium complex species. Microbiology. 2008. 154:3715–3723.
Article22. Kim BJ, Lee KH, Park BN, Kim SJ, Bai GH, Kim SJ, et al. Differentiation of mycobacterial species by PCR-restriction analysis of DNA (342 base pairs) of the RNA polymerase gene (rpoB). J Clin Microbiol. 2001. 39:2102–2109.
Article23. Lee H, Park HJ, Cho SN, Bai GH, Kim SJ. Species identification of mycobacteria by PCR-restriction fragment length polymorphism of the rpoB gene. J Clin Microbiol. 2000. 38:2966–2971.
Article24. Suffys PN, da Silva Rocha A, de Oliveira M, Campos CE, Barreto AM, Portaels F, et al. Rapid identification of Mycobacteria to the species level using INNO-LiPA Mycobacteria, a reverse hybridization assay. J Clin Microbiol. 2001. 39:4477–4482.
Article25. Xiong L, Kong F, Yang Y, Cheng J, Gilbert GL. Use of PCR and reverse line blot hybridization macroarray based on 16S-23S rRNA gene internal transcribed spacer sequences for rapid identification of 34 mycobacterium species. J Clin Microbiol. 2006. 44:3544–3550.
Article26. Phillips RO, Sarfo FS, Osei-Sarpong F, Boateng A, Tetteh I, Lartey A, et al. Sensitivity of PCR for M. ulcerans on Fine Needle Aspirates for Diagnosis of Buruli ulcer. J Clin Microbiol. 2009. 47:924–926.
Article27. Stinear T, Ross BC, Davies JK, Marino L, Robins-Browne RM, Oppedisano F, et al. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J Clin Microbiol. 1999. 37:1018–1023.
Article28. Koivula T, Cristea-Fernström M, Chryssanthou E, Petrini B, Källenius G. Genetic diversity in clinical isolates of Mycobacterium avium complex from Guinea-Bissau, West Africa. Microbes Infec. 2004. 6:1320–1325.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Identification of Mycobacterium Species by Multiplex PCR-Restriction Fragment Length Polymorphism Assay
- Multiplex TaqMan qPCR Assay for Detection, Identification, and Quantification of Three Sclerotinia Species
- Identification of Candida Species by Multiplex Polymerase Chain Reaction
- Multiplex Real-time Polymerase Chain Reaction Assays for Simultaneous Detection of
Vibrio cholerae ,Vibrio parahaemolyticus , andVibrio vulnificus - Identification of Mycobacteria using High Performance Liquid Chromatography in Clinical Specimens