Healthc Inform Res.
2014 Jan;20(1):61-68. 10.4258/hir.2014.20.1.61.
Online Learning for Classification of Alzheimer Disease based on Cortical Thickness and Hippocampal Shape Analysis
- Affiliations
-
- 1Department of Biomedical Engineering, Korea University, Seoul, Korea. jkseong@korea.ac.kr
- 2Department of Computer and Radio Communications Engineering, Korea University, Seoul, Korea.
- 3Seoul National University Biomedical informatics (SNUBI), Division of Biomedical Informatics, Seoul National University College of Medicine, Seoul, Korea.
- 4Systems Biomedical Informatics Research Center, Seoul National University, Seoul, Korea.
- 5Department of Neuropsychiatry, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seongnam, Korea.
- KMID: 2166740
- DOI: http://doi.org/10.4258/hir.2014.20.1.61
Abstract
OBJECTIVES
Mobile healthcare applications are becoming a growing trend. Also, the prevalence of dementia in modern society is showing a steady growing trend. Among degenerative brain diseases that cause dementia, Alzheimer disease (AD) is the most common. The purpose of this study was to identify AD patients using magnetic resonance imaging in the mobile environment.
METHODS
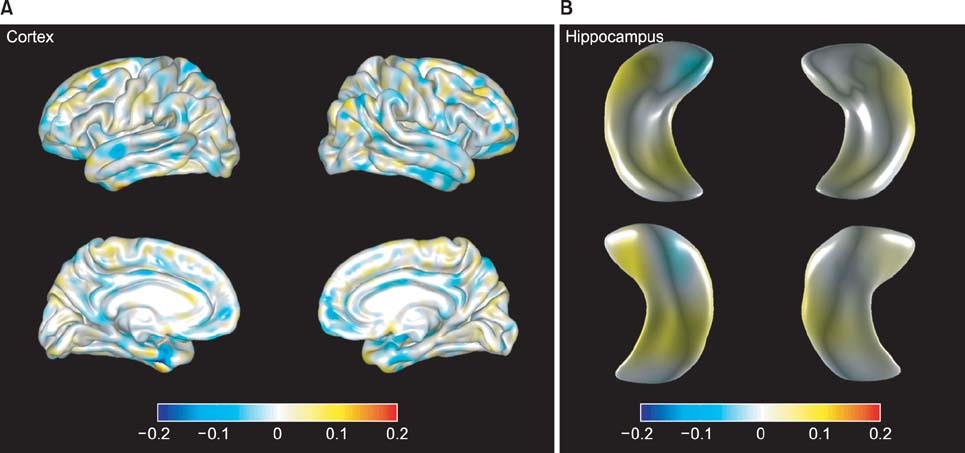
We propose an incremental classification for mobile healthcare systems. Our classification method is based on incremental learning for AD diagnosis and AD prediction using the cortical thickness data and hippocampus shape. We constructed a classifier based on principal component analysis and linear discriminant analysis. We performed initial learning and mobile subject classification. Initial learning is the group learning part in our server. Our smartphone agent implements the mobile classification and shows various results.
RESULTS
With use of cortical thickness data analysis alone, the discrimination accuracy was 87.33% (sensitivity 96.49% and specificity 64.33%). When cortical thickness data and hippocampal shape were analyzed together, the achieved accuracy was 87.52% (sensitivity 96.79% and specificity 63.24%).
CONCLUSIONS
In this paper, we presented a classification method based on online learning for AD diagnosis by employing both cortical thickness data and hippocampal shape analysis data. Our method was implemented on smartphone devices and discriminated AD patients for normal group.
Keyword
MeSH Terms
Figure
Reference
-
1. Martinez-Perez B, de la Torre-Diez I, Lopez-Coronado M. Mobile health applications for the most prevalent conditions by the World Health Organization: review and analysis. J Med Internet Res. 2013; 15(6):e120.
Article2. Boulos MN, Wheeler S, Tavares C, Jones R. How smartphones are changing the face of mobile and participatory healthcare: an overview, with example from eCAALYX. Biomed Eng Online. 2011; 10:24.
Article3. Jack CR Jr. Alzheimer disease: new concepts on its neurobiology and the clinical role imaging will play. Radiology. 2012; 263(2):344–361.
Article4. Bain LJ, Jedrziewski K, Morrison-Bogorad M, Albert M, Cotman C, Hendrie H, et al. Healthy brain aging: a meeting report from the Sylvan M. Cohen Annual Retreat of the University of Pennsylvania Institute on Aging. Alzheimers Dement. 2008; 4(6):443–446.
Article5. Jack CR Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010; 9(1):119–128.
Article6. Desikan RS, Cabral HJ, Hess CP, Dillon WP, Glastonbury CM, Weiner MW, et al. Automated MRI measures identify individuals with mild cognitive impairment and Alzheimer's disease. Brain. 2009; 132(Pt 8):2048–2057.
Article7. Querbes O, Aubry F, Pariente J, Lotterie JA, Demonet JF, Duret V, et al. Early diagnosis of Alzheimer's disease using cortical thickness: impact of cognitive reserve. Brain. 2009; 132(Pt 8):2036–2047.
Article8. Colliot O, Chetelat G, Chupin M, Desgranges B, Magnin B, Benali H, et al. Discrimination between Alzheimer disease, mild cognitive impairment, and normal aging by using automated segmentation of the hippocampus. Radiology. 2008; 248(1):194–201.
Article9. Gerardin E, Chetelat G, Chupin M, Cuingnet R, Desgranges B, Kim HS, et al. Multidimensional classification of hippocampal shape features discriminates Alzheimer's disease and mild cognitive impairment from normal aging. Neuroimage. 2009; 47(4):1476–1486.
Article10. Hall PM, Marchall D, Martin RR. Incremental eigenanalysis for classification. Cardiff, UK: Department of Computer Science, University of Wales;1998.11. Pang S, Ozawa S, Kasabov N. Incremental linear discriminant analysis for classification of data streams. IEEE Trans Syst Man Cybern B Cybern. 2005; 35(5):905–914.
Article12. Cho Y, Seong JK, Jeong Y, Shin SY. Alzheimer's Disease Neuroimaging Initiative. Individual subject classification for Alzheimer's disease based on incremental learning using a spatial frequency representation of cortical thickness data. Neuroimage. 2012; 59(3):2217–2230.
Article13. Zhao W, Chellappa R, Phillips PJ, Rosenfeld A. Face recognition: a literature survey. ACM Comput Surv. 2003; 35(4):399–458.14. Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage. 1999; 9(2):179–194.15. Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis: II. Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999; 9(2):195–207.16. Segonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, et al. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004; 22(3):1060–1075.
Article17. Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998; 17(1):87–97.
Article18. Fischl B, Liu A, Dale AM. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans Med Imaging. 2001; 20(1):70–80.
Article19. Segonne F, Pacheco J, Fischl B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans Med Imaging. 2007; 26(4):518–529.
Article20. Belhumeur PN, Hespanha JP, Kriegman D. Eigenfaces vs. fisherfaces: recognition using class specific linear projection. IEEE Trans Pattern Anal Mach Intell. 1997; 19(7):711–720.
Article21. Liu C, Wechsler H. Robust coding schemes for indexing and retrieval from large face databases. IEEE Trans Image Process. 2000; 9(1):132–137.
Article22. Yu H, Yang J. A direct LDA algorithm for high-dimensional data: with application to face recognition. Pattern Recognit. 2001; 34(10):2067–2070.
Article23. Balakrishnama S, Ganapathiraju A. Linear discriminant analysis: a brief tutorial. Mississippi State (MS): Institute for Signal and information Processing, Mississippi State University;1998.24. Lim J, Ross D, Lin RS, Yang MH. Incremental learning for visual tracking. Advances in neural information processing systems 17. Cambridge (MA): MIT Press;2004. p. 793–800.25. Hall PM, Marshall D, Martin RR. Adding and subtracting eigenspaces with eigenvalue decomposition and singular value decomposition. Image Vis Comput. 2002; 20(13):1009–1016.
Article26. Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehericy S, Habert MO, et al. Automatic classification of patients with Alzheimer's disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage. 2011; 56(2):766–781.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Neuroimaging Markers for Alzheimer's Disease and Mild Cognitive Impairment in Alzheimer's Disease Neuroimaging Initiative (ADNI)
- Changes in the Hippocampal Volume and Shape in Early-Onset Mild Cognitive Impairment
- Hippocampal Atrophy and Psychotic Symptoms in Patients with Alzheimer's Disease
- Future directions of online learning environment design at medical schools: a transition towards a post-pandemic context
- The influence of learning presence and self-directed learning competency of nursing students on learning satisfaction in major subjects for online distance learning