TCF4-Targeting miR-124 is Differentially Expressed amongst Dendritic Cell Subsets
- Affiliations
-
- 1Laboratory of Immunology, Severance Biomedical Science Institute, Yonsei University College of Medicine, Seoul 03722, Korea. ChaeGyu@yuhs.ac
- 2Brain Korea 21 PLUS Project for Medical Science, Severance Biomedical Science Institute, Yonsei University College of Medicine, Seoul 03722, Korea.
- 3Institute for Bio-Medical Convergence, College of Medicine, Catholic Kwandong University, Gangneung 25601, Korea.
- KMID: 2154812
- DOI: http://doi.org/10.4110/in.2016.16.1.61
Abstract
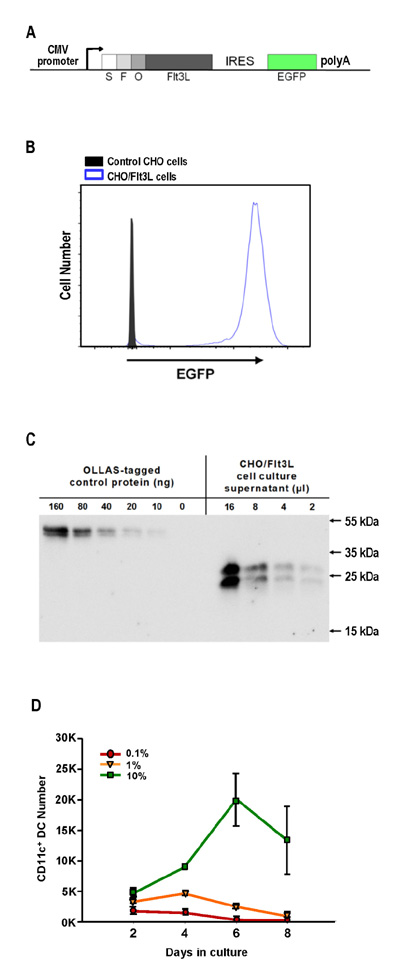
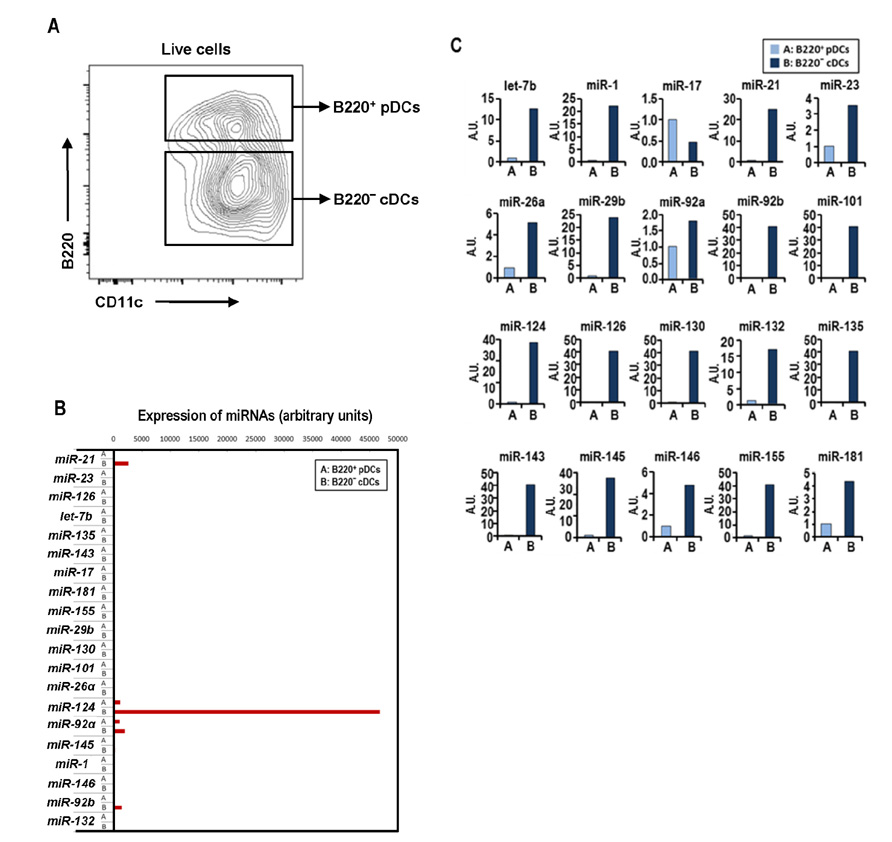
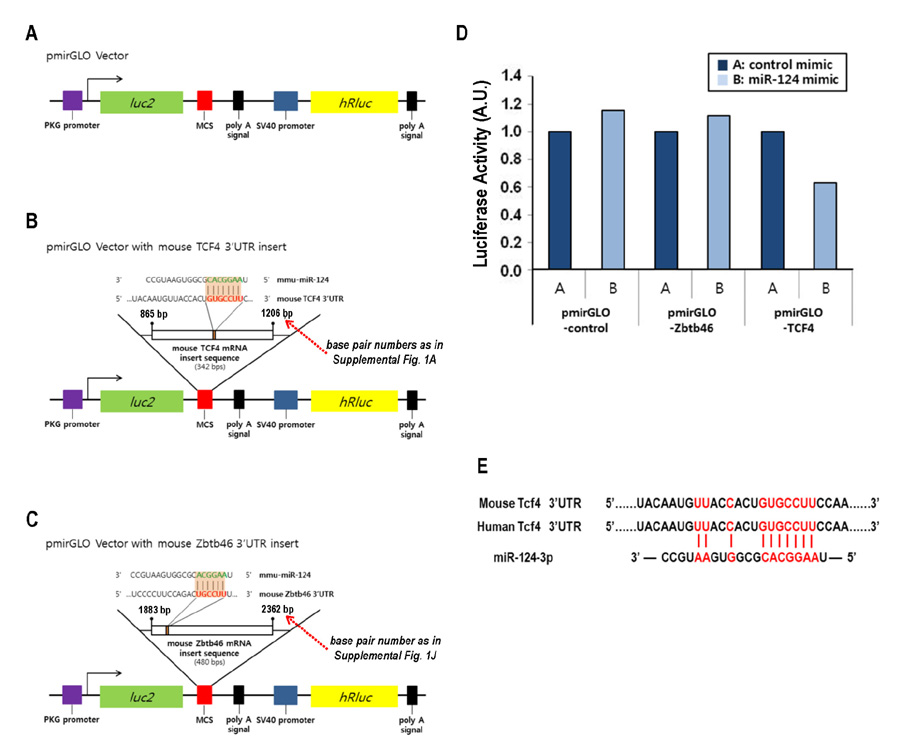
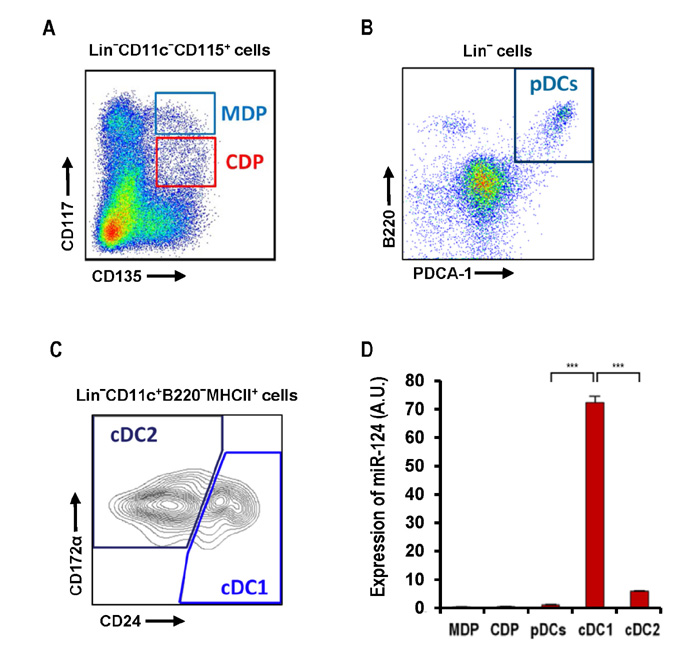
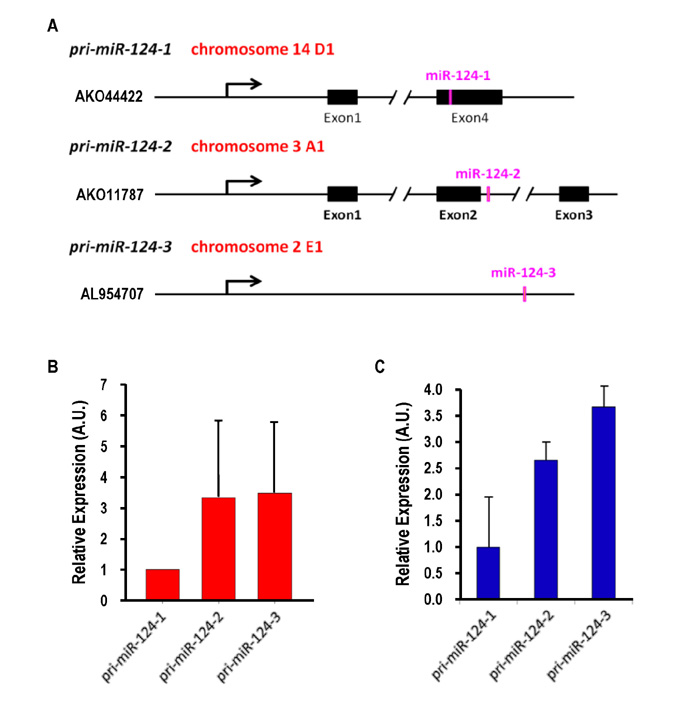
- Dendritic cells (DCs) are professional antigen-presenting cells that sample their environment and present antigens to naive T lymphocytes for the subsequent antigen-specific immune responses. DCs exist in a range of distinct subpopulations including plasmacytoid DCs (pDCs) and classical DCs (cDCs), with the latter consisting of the cDC1 and cDC2 lineages. Although the roles of DC-specific transcription factors across the DC subsets have become understood, the posttranscriptional mechanisms that regulate DC development are yet to be elucidated. MicroRNAs (miRNAs) are pivotal posttranscriptional regulators of gene expression in a myriad of biological processes, but their contribution to the immune system is just beginning to surface. In this study, our in-house probe collection was screened to identify miRNAs possibly involved in DC development and function by targeting the transcripts of relevant mouse transcription factors. Examination of DC subsets from the culture of mouse bone marrow with Flt3 ligand identified high expression of miR-124 which was able to target the transcript of TCF4, a transcription factor critical for the development and homeostasis of pDCs. Further expression profiling of mouse DC subsets isolated from in vitro culture as well as via ex vivo purification demonstrated that miR-124 was outstandingly expressed in CD24+ cDC1 cells compared to in pDCs and CD172alpha+ cDC2 cells. These results imply that miR-124 is likely involved in the processes of DC subset development by posttranscriptional regulation of a transcription factor(s).
MeSH Terms
Figure
Cited by 3 articles
-
Two Distinct Subsets Are Identified from the Peritoneal Myeloid Mononuclear Cells Expressing both CD11c and CD115
Moah Sohn, Hye Young Na, Seul Hye Ryu, Wanho Choi, Hyunju In, Hyun Soo Shin, Ji Soo Park, Dahee Shim, Sung Jae Shin, Chae Gyu Park
Immune Netw. 2019;19(3):. doi: 10.4110/in.2019.19.e15.Extended Culture of Bone Marrow with Granulocyte Macrophage-Colony Stimulating Factor Generates Immunosuppressive Cells
Hye Young Na, Moah Sohn, Seul Hye Ryu, Wanho Choi, Hyunju In, Hyun Soo Shin, Chae Gyu Park
Immune Netw. 2018;18(2):e16. doi: 10.4110/in.2018.18.e16.Extended Culture of Bone Marrow with Granulocyte Macrophage-Colony Stimulating Factor Generates Immunosuppressive Cells
Hye Young Na, Moah Sohn, Seul Hye Ryu, Wanho Choi, Hyunju In, Hyun Soo Shin, Chae Gyu Park
Immune Netw. 2018;18(2):. doi: 10.4110/in.2018.18.e16.
Reference
-
1. Steinman RM. Decisions about dendritic cells: past, present, and future. Annu Rev Immunol. 2012; 30:1–22.
Article2. Park CG. Vaccine strategies utilizing C-type lectin receptors on dendritic cells in vivo. Clin Exp Vaccine Res. 2014; 3:149–154.
Article3. Steinman RM, Idoyaga J. Features of the dendritic cell lineage. Immunol Rev. 2010; 234:5–17.
Article4. Kingston D, Schmid MA, Onai N, Obata-Onai A, Baumjohann D, Manz MG. The concerted action of GM-CSF and Flt3-ligand on in vivo dendritic cell homeostasis. Blood. 2009; 114:835–843.
Article5. Onai N, Kurabayashi K, Hosoi-Amaike M, Toyama-Sorimachi N, Matsushima K, Inaba K, Ohteki T. A clonogenic progenitor with prominent plasmacytoid dendritic cell developmental potential. Immunity. 2013; 38:943–957.
Article6. Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, Kc W, Kretzer NM, Briseno CG, Durai V, Bagadia P, Haldar M, Schonheit J, Rosenbauer F, Murphy TL, Murphy KM. Batf3 maintains autoactivation of Irf8 for commitment of a CD8alpha(+) conventional DC clonogenic progenitor. Nat Immunol. 2015; 16:708–717.
Article7. Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HR, Schreuder J, Lum J, Malleret B, Zhang S, Larbi A, Zolezzi F, Renia L, Poidinger M, Naik S, Newell EW, Robson P, Ginhoux F. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol. 2015; 16:718–728.
Article8. Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010; 327:656–661.
Article9. Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nat Immunol. 2012; 13:1145–1154.
Article10. Murphy KM. Transcriptional control of dendritic cell development. Adv Immunol. 2013; 120:239–267.
Article11. Esashi E, Wang YH, Perng O, Qin XF, Liu YJ, Watowich SS. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity. 2008; 28:509–520.
Article12. Li Z, Rana TM. Therapeutic targeting of micro-RNAs: current status and future challenges. Nat Rev Drug Discov. 2014; 13:622–638.
Article13. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014; 15:509–524.
Article14. Johanson TM, Skinner JP, Kumar A, Zhan Y, Lew AM, Chong MM. The role of microRNAs in lymphopoiesis. Int J Hematol. 2014; 100:246–253.
Article15. Smyth LA, Boardman DA, Tung SL, Lechler R, Lombardi G. MicroRNAs affect dendritic cell function and phenotype. Immunology. 2015; 144:197–205.
Article16. Mildner A, Chapnik E, Manor O, Yona S, Kim KW, Aychek T, Varol D, Beck G, Itzhaki ZB, Feldmesser E, Amit I, Hornstein E, Jung S. Mononuclear phagocyte miRNome analysis identifies miR-142 as critical regulator of murine dendritic cell homeostasis. Blood. 2013; 121:1016–1027.
Article17. Karrich JJ, Jachimowski LC, Libouban M, Iyer A, Brandwijk K, Taanman-Kueter EW, Nagasawa M, de Jong EC, Uittenbogaart CH, Blom B. MicroRNA-146a regulates survival and maturation of human plasmacytoid dendritic cells. Blood. 2013; 122:3001–3009.
Article18. Su X, Qian C, Zhang Q, Hou J, Gu Y, Han Y, Chen Y, Jiang M, Cao X. miRNomes of haematopoietic stem cells and dendritic cells identify miR-30b as a regulator of Notch1. Nat Commun. 2013; 4:2903.
Article19. Agudo J, Ruzo A, Tung N, Salmon H, Leboeuf M, Hashimoto D, Becker C, Garrett-Sinha LA, Baccarini A, Merad M, Brown BD. The miR-126-VEGFR2 axis controls the innate response to pathogen-associated nucleic acids. Nat Immunol. 2014; 15:54–62.
Article20. Park H, Huang X, Lu C, Cairo MS, Zhou X. MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J Biol Chem. 2015; 290:2831–2841.
Article21. Johanson TM, Cmero M, Wettenhall J, Lew AM, Zhan Y, Chong MM. A microRNA expression atlas of mouse dendritic cell development. Immunol Cell Biol. 2015; 93:480–485.
Article22. Park SH, Cheong C, Idoyaga J, Kim JY, Choi JH, Do Y, Lee H, Jo JH, Oh YS, Im W, Steinman RM, Park CG. Generation and application of new rat monoclonal antibodies against synthetic FLAG and OLLAS tags for improved immunodetection. J Immunol Methods. 2008; 331:27–38.
Article23. Inaba K, Swiggard WJ, Steinman RM, Romani N, Schuler G, Brinster C. Isolation of dendritic cells. Curr Protoc Immunol. 2009; Chapter 3:Unit 3.7.
Article24. Real FM, Sekido R, Lupianez DG, Lovell-Badge R, Jimenez R, Burgos M. A microRNA (mmu-miR-124) prevents Sox9 expression in developing mouse ovarian cells. Biol Reprod. 2013; 89:78.25. Darrasse-Jeze G, Deroubaix S, Mouquet H, Victora GD, Eisenreich T, Yao KH, Masilamani RF, Dustin ML, Rudensky A, Liu K, Nussenzweig MC. Feedback control of regulatory T cell homeostasis by dendritic cells in vivo. J Exp Med. 2009; 206:1853–1862.26. Choi JH, Cheong C, Dandamudi DB, Park CG, Rodriguez A, Mehandru S, Velinzon K, Jung IH, Yoo JY, Oh GT, Steinman RM. Flt3 signaling-dependent dendritic cells protect against atherosclerosis. Immunity. 2011; 35:819–831.
Article27. Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015; 12:697.
Article28. Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, Holmberg D, Zweier C, den Hollander NS, Kant SG, Holter W, Rauch A, Zhuang Y, Reizis B. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008; 135:37–48.
Article29. Naik SH, Proietto AI, Wilson NS, Dakic A, Schnorrer P, Fuchsberger M, Lahoud MH, O'Keeffe M, Shao QX, Chen WF, Villadangos JA, Shortman K, Wu L. Cutting edge: generation of splenic CD8+ and CD8− dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol. 2005; 174:6592–6597.
Article30. Naik SH, Sathe P, Park HY, Metcalf D, Proietto AI, Dakic A, Carotta S, O'Keeffe M, Bahlo M, Papenfuss A, Kwak JY, Wu L, Shortman K. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007; 8:1217–1226.
Article31. Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, Chu FF, Randolph GJ, Rudensky AY, Nussenzweig M. In vivo analysis of dendritic cell development and homeostasis. Science. 2009; 324:392–397.
Article32. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73.
Article33. Qiu S, Feng Y, LeSage G, Zhang Y, Stuart C, He L, Li Y, Caudle Y, Peng Y, Yin D. Chronic morphine-induced microRNA-124 promotes microglial immunosuppression by modulating P65 and TRAF6. J Immunol. 2015; 194:1021–1030.
Article34. Baudet ML, Zivraj KH, breu-Goodger C, Muldal A, Armisen J, Blenkiron C, Goldstein LD, Miska EA, Holt CE. miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat Neurosci. 2012; 15:29–38.
Article35. Sonntag KC, Woo TU, Krichevsky AM. Converging miRNA functions in diverse brain disorders: a case for miR-124 and miR-126. Exp Neurol. 2012; 235:427–435.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- MiR-103a-3p Contributes to the Progression of Colorectal Cancer by Regulating GREM2 Expression
- Differential MicroRNA Expression between EGFR T790M and L858R Mutated Lung Cancer
- Regulation of Th2 Cell Immunity by Dendritic Cells
- Helper T Cell Polarizing Through Dendritic Cells
- Circular RNA-ZFR Inhibited Cell Proliferation and Promoted Apoptosis in Gastric Cancer by Sponging miR-130a/miR-107 and Modulating PTEN