Korean J Clin Microbiol.
2011 Jun;14(2):41-47. 10.5145/KJCM.2011.14.2.41.
Characteristics of Molecular Strain Typing of Mycobacterium tuberculosis Isolated from Korea
- Affiliations
-
- 1Department of Laboratory Medicine, School of Medicine, Pusan National University, Busan, Korea.
- 2Department of Thoracic and Cardiovascular Surgery, School of Medicine, Pusan National University, Busan, Korea. domini@pnu.edu
- KMID: 2089813
- DOI: http://doi.org/10.5145/KJCM.2011.14.2.41
Abstract
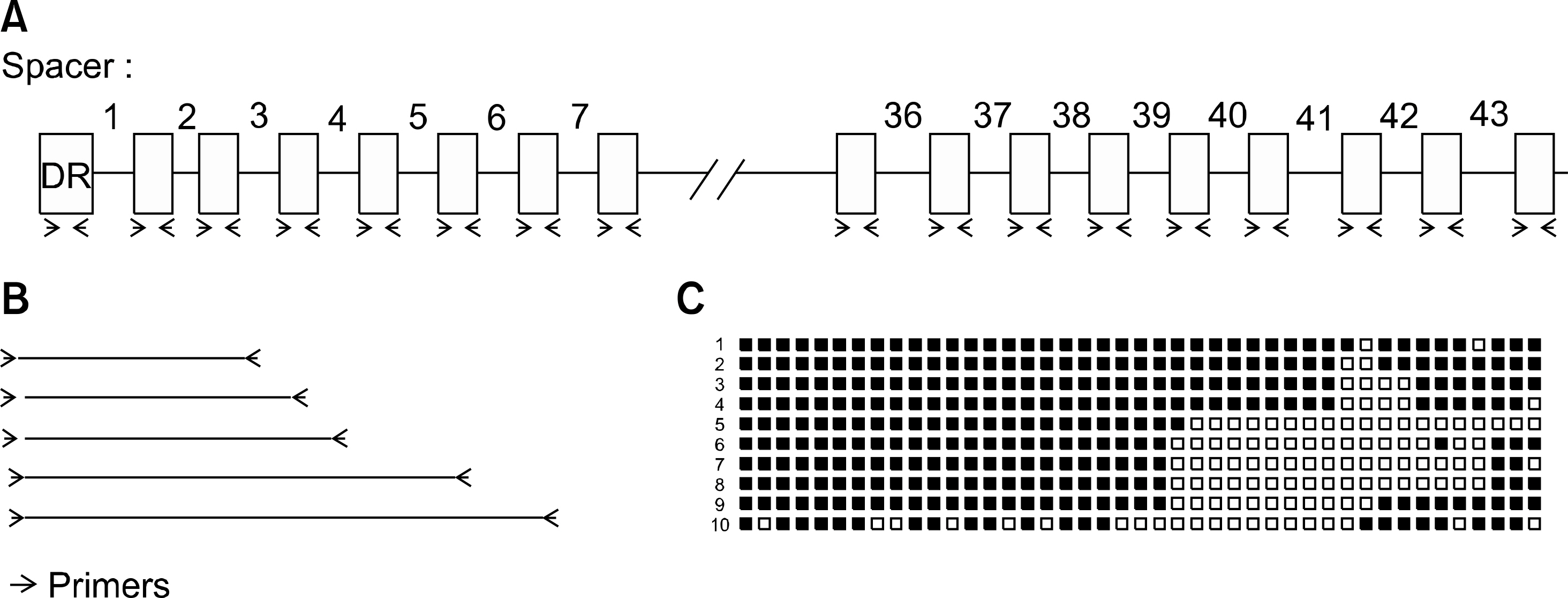
- Molecular strain typing of Mycobacterium tuberculosis is important for the detection of outbreaks of tuberculosis and laboratory cross contamination, as well as the differentiation between re-infection and reactivation of tuberculosis. In the present review, the authors investigated the currently available typing methods for M. tuberculosis and the current status of strain distribution in Korea. IS6110-restriction fragment length polymorphism (RFLP), which is considered a standard method, is based on numbers and positions of the insertion sequence, IS6110. The method has an excellent discriminatory power with a considerable amount of worldwide data, although it is time-consuming and labor-intensive. Spoligotyping is based on the presence or absence of spacer sequences between direct repeat (DR) regions. PCR amplification allows for the possibility of application in the early suspicious stage. The data can be easily digitized; however, it shows identical profiles in Beijing family strains. Mycobacterial interspersed repetitive unit-variable number of tandem repeat (MIRU-VNTR) is another PCR-based genotyping method with a good discrimination power whose data can also be easily digitized. In Korea, the prevalence of Beijing family strains have been as high as 80 to 87%.
MeSH Terms
Figure
Reference
-
1. Hong YP, Kim SJ, Lew WJ, Lee EK, Han YC. The seventh nationwide tuberculosis prevalence survey in Korea, 1995. Int J Tuberc Lung Dis. 1998; 2:27–36.2. World Health Organization. Tuberculosis control in the Western Pacific region: 2008 Report;. 2008. http://www.medcastle.com/tuberculosis-control-in-the-western-pacific-region-2008-report.html.3. Aristimuño L, Armengol R, Cebollada A, España M, Guilarte A, Lafoz C, et al. Molecular characterisation of Mycobacterium tuberculosis isolates in the First National Survey of Anti-tuberculosis Drug Resistance from Venezuela. BMC Microbiol. 2006; 6:90.
Article4. Kim SJ, Bai GH, Lee H, Kim HJ, Lew WJ, Park YK, et al. Transmission of Mycobacterium tuberculosis among high school students in Korea. Int J Tuberc Lung Dis. 2001; 5:824–30.5. Kan B, Berggren I, Ghebremichael S, Bennet R, Bruchfeld J, Chryssanthou E, et al. Extensive transmission of an isoniazid-resistant strain of Mycobacterium tuberculosis in Sweden. Int J Tuberc Lung Dis. 2008; 12:199–204.6. Tostmann A, Kik SV, Kalisvaart NA, Sebek MM, Verver S, Boeree MJ, et al. Tuberculosis transmission by patients with smear-negative pulmonary tuberculosis in a large cohort in the Netherlands. Clin Infect Dis. 2008; 47:1135–42.
Article7. van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993; 31:406–9.
Article8. Gómez Marín JE, Rigouts L, Villegas Londoño LE, Portaels F. Analysis of restriction fragment length polymorphism (RFLP) in epidemiology of tuberculosis. Bol Oficina Sanit Panam. 1995; 119:1–10.9. van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991; 29:2578–86.
Article10. Glynn JR, Whiteley J, Bifani PJ, Kremer K, van Soolingen D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg Infect Dis. 2002; 8:843–9.11. European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg Infect Dis. 2006; 12:736–43.12. Rhee JT, Tanaka MM, Behr MA, Agasino CB, Paz EA, Hopewell PC, et al. Use of multiple markers in population-based molecular epidemiologic studies of tuberculosis. Int J Tuberc Lung Dis. 2000; 4:1111–9.13. Asgharzadeh M, Khakpour M, Salehi TZ, Kafil HS. Use of mycobacterial interspersed repetitive unit-variable-number tandem repeat typing to study Mycobacterium tuberculosis isolates from East Azarbaijan province of Iran. Pak J Biol Sci. 2007; 10:3769–77.14. Lee AS, Tang LL, Lim IH, Bellamy R, Wong SY. Discrimination of single-copy IS6110 DNA fingerprints of Mycobacterium tuberculosis isolates by high-resolution minisatellite-based typing. J Clin Microbiol. 2002; 40:657–9.15. Mazars E, Lesjean S, Banuls AL, Gilbert M, Vincent V, Gicquel B, et al. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc Natl Acad Sci USA. 2001; 98:1901–6.
Article16. Radhakrishnan I, K MY, Kumar RA, Mundayoor S. Implications of low frequency of IS6110 in fingerprinting field isolates of Mycobacterium tuberculosis from Kerala, India. J Clin Microbiol. 2001; 39:1683.17. Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997; 35:907–14.
Article18. Gori A, Bandera A, Marchetti G, Degli Esposti A, Catozzi L, Nardi GP, et al. Spoligotyping and Mycobacterium tuberculosis. Emerg Infect Dis. 2005; 11:1242–8.19. Gori A, Esposti AD, Bandera A, Mezzetti M, Sola C, Marchetti G, et al. Comparison between spoligotyping and IS6110 restriction fragment length polymorphisms in molecular genotyping analysis of Mycobacterium tuberculosis strains. Mol Cell Probes. 2005; 19:236–44.
Article20. Kwara A, Schiro R, Cowan LS, Hyslop NE, Wiser MF, Roahen Harrison S, et al. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J Clin Microbiol. 2003; 41:2683–5.21. Filliol I, Ferdinand S, Sola C, Thonnon J, Rastogi N. Spoligotyping and IS6110-RFLP typing of Mycobacterium tuberculosis from French Guiana: a comparison of results with international databases underlines interregional transmission from neighboring countries. Res Microbiol. 2002; 153:81–8.
Article22. Filliol I, Driscoll JR, Van Soolingen D, Kreiswirth BN, Kremer K, Valétudie G, et al. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg Infect Dis. 2002; 8:1347–9.
Article23. Diaz R, Kremer K, de Haas PE, Gomez RI, Marrero A, Valdivia JA, et al. Molecular epidemiology of tuberculosis in Cuba outside of Havana, July 1994–June 1995: utility of spoligotyping versus IS6110 restriction fragment length polymorphism. Int J Tuberc Lung Dis. 1998; 2:743–50.24. Doroudchi M, Kremer K, Basiri EA, Kadivar MR, Van Soolingen D, Ghaderi AA. IS6110-RFLP and spoligotyping of Mycobacterium tuberculosis isolates in Iran. Scand J Infect Dis. 2000; 32:663–8.25. Song EJ, Jeong HJ, Lee SM, Kim CM, Song ES, Park YK, et al. A DNA chip-based spoligotyping method for the strain identification of Mycobacterium tuberculosis isolates. J Microbiol Methods. 2007; 68:430–3.
Article26. Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006; 44:4498–510.27. Oelemann MC, Diel R, Vatin V, Haas W, Rüsch-Gerdes S, Locht C, et al. Assessment of an optimized mycobacterial interspersed repetitive- unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J Clin Microbiol. 2007; 45:691–7.
Article28. Bouakaze C, Keyser C, de Martino SJ, Sougakoff W, Veziris N, Dabernat H, et al. Identification and genotyping of Mycobacterium tuberculosis complex species by use of a SNaPshot Minise-quencing-based assay. J Clin Microbiol. 2010; 48:1758–66.29. Mahasirimongkol S, Yanai H, Nishida N, Ridruechai C, Matsushita I, Ohashi J, et al. Genome-wide SNP-based linkage analysis of tuberculosis in Thais. Genes Immun. 2009; 10:77–83.
Article30. van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, Portaels F, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol. 1995; 33:3234–8.
Article31. Park YK, Bai GH, Kim SJ. Restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolated from countries in the western pacific region. J Clin Microbiol. 2000; 38:191–7.32. Kang HY, Wada T, Iwamoto T, Maeda S, Murase Y, Kato S, et al. Phylogeographical particularity of the Mycobacterium tuberculosis Beijing family in South Korea based on international comparison with surrounding countries. J Med Microbiol. 2010; 59:1191–7.
Article33. Choi GE, Jang MH, Song EJ, Jeong SH, Kim JS, Lee WG, et al. IS6110-restriction fragment length polymorphism and spoligotyping analysis of Mycobacterium tuberculosis clinical isolates for investigating epidemiologic distribution in Korea. J Korean Med Sci. 2010; 25:1716–21.34. Shamputa IC, Lee J, Allix-Béguec C, Cho EJ, Lee JI, Rajan V, et al. Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary care tuberculosis hospital in South Korea. J Clin Microbiol. 2010; 48:387–94.35. Jeon CH, Lee SC, Sohn JH, Ahn WS. Restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolated from Taegu. Korean J Clin Pathol. 1999; 19:581–6.36. Park YK, Kang HY, Lim JG, Ha JS, Cho JO, Lee KC, et al. Analysis of DNA fingerprints of Mycobacterium tuberculosis isolates from patients registered at health center in Gyeonggi Province in 2004. Tuberc Respir Dis. 2006; 60:290–6.
Article37. Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 2002; 10:45–52.
Article38. Qian L, Abe C, Lin TP, Yu MC, Cho SN, Wang S, et al. rpoB genotypes of Mycobacterium tuberculosis Beijing family isolates from East Asian countries. J Clin Microbiol. 2002; 40:1091–4.39. Park YK, Shin S, Ryu S, Cho SN, Koh WJ, Kwon OJ, et al. Comparison of drug resistance genotypes between Beijing and non-Beijing family strains of Mycobacterium tuberculosis in Korea. J Microbiol Methods. 2005; 63:165–72.
Article40. Chang CL, Kim HH, Son HC, Park SS, Lee MK, Park SK, et al. False-positive growth of Mycobacterium tuberculosis attributable to laboratory contamination confirmed by restriction fragment length polymorphism analysis. Int J Tuberc Lung Dis. 2001; 5:861–7.41. Yun KW, Song EJ, Choi GE, Hwang IK, Lee EY, Chang CL. Strain typing of Mycobacterium tuberculosis isolates from Korea by mycobacterial interspersed repetitive units-variable number of tandem repeats. Korean J Lab Med. 2009; 29:314–9.
Article42. Hanekom M, van der Spuy GD, Gey van Pittius NC, McEvoy CR, Hoek KG, Ndabambi SL, et al. Discordance between mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing and IS6110 restriction fragment length polymorphism genotyping for analysis of Mycobacterium tuberculosis Beijing strains in a setting of high incidence of tuberculosis. J Clin Microbiol. 2008; 46:3338–45.43. Filliol I, Motiwala AS, Cavatore M, Qi W, Hazbón MH, Bobadilla del Valle M, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006; 188:759–72.44. Choi GE, Jang MH, Cho HJ, Lee SM, Yi J, Lee EY, et al. Application of single-nucleotide polymorphism and mycobacterial interspersed repetitive units-variable number of tandem repeats analyses to clinical Mycobacterium tuberculosis isolates from Korea. Korean J Lab Med. 2011; 31:37–43.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular Strain Typing of Mycobacterium tuberculosis: a Review of Frequently Used Methods
- Optimal Combination of VNTR Typing for Discrimination of Isolated Mycobacterium tuberculosis in Korea
- Molecular fingerprinting of clinical isolates of Mycobacterium bovis and Mycobacterium tuberculosis from India by restriction fragment length polymorphism (RFLP)
- Application of Infrequent-Restriction-Site Amplification for Genotyping of Mycobacterium tuberculosis and non-tuberculous Mycobacterium
- Molecular and genomic features of Mycobacterium bovis strain 1595 isolated from Korean cattle