J Breast Cancer.
2013 Dec;16(4):366-371. 10.4048/jbc.2013.16.4.366.
Case-Control Study on the Fibroblast Growth Factor Receptor 2 Gene Polymorphisms Associated with Breast Cancer in Chinese Han Women
- Affiliations
-
- 1Department of Center for Reproductive Medicine, General Hospital, Ningxia Medical University, Ningxia, China.
- 2Key Laboratory of Fertility Preservation and Maintenance, Ningxia Medical University, Ministry of Education, Ningxia, China. hyjiao1602@hotmail.com
- 3Department of Clinical Laboratory, Ningxiang Hospital, Hunan, China.
- 4Department of Surgical Oncology, General Hospital, Ningxia Medical University, Ningxia, China.
- 5Department of Clinical Laboratory, General Hospital, Ningxia Medical University, Ningxia, China.
- 6Department of Medical Genetics and Cell Biology, Ningxia Medical University, Ningxia, China.
- KMID: 1976373
- DOI: http://doi.org/10.4048/jbc.2013.16.4.366
Abstract
- PURPOSE
Genetic variation in fibroblast growth factor receptor 2 (FGFR2) is a newly described risk factor for breast cancer. This study aimed to evaluate the association of four single nucleotide polymorphisms (SNPs) in FGFR2 with breast cancer in Han Chinese women.
METHODS
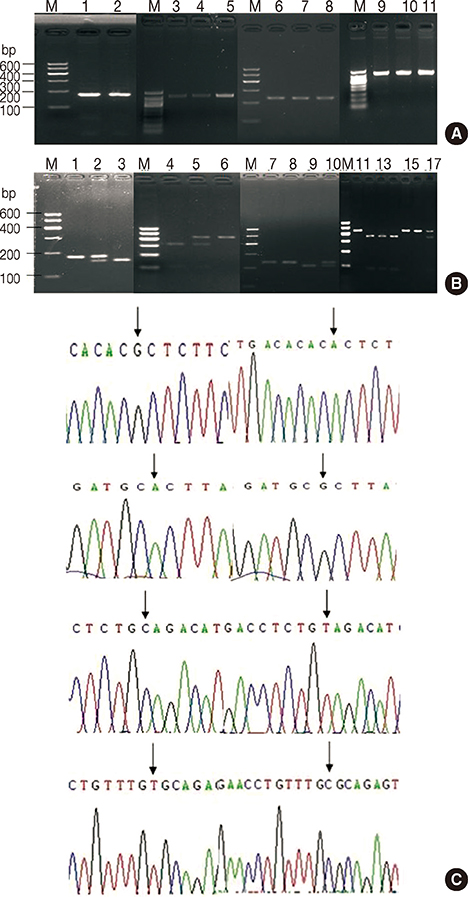
Two hundred three women with breast cancer and 200 breast cancer-free age-matched controls were selected. Four SNPs (rs2981579, rs1219648, rs2420946, and rs2981582) and their haplotypes were analyzed to test for their association with breast cancer susceptibility. The presence of the four FGFR2 SNPs was determined by polymerase chain reaction-restriction fragment length polymorphism analysis.
RESULTS
A statistically significant difference was observed in the frequency of rs2981582 in the FGFR2 gene (p<0.05) between case and control groups. In subjects stratified by menopausal status, rs2981582 TT, rs2420946 AA, and rs1219648 CC were significantly associated with the risk of breast cancer in postmenopausal subjects, but no significant associations between these four SNPs and the risk of breast cancer were identified in premenopausal subjects. Further, there was no significant association between hormone receptor status (estrogen receptor and progesterone receptor) and breast cancer risk. Six common (> 3%) haplotypes were identified. Three of these haplotypes, CGTC (odds ratio [OR], 0.613; 95% confidence interval [CI], 0.457-0.82; p=0.001), TGTC (OR, 6.561; 95% CI, 2.064-20.854; p<0.001), and CATC (OR, 12.645; 95% CI, 1.742-91.799; p=0.001) were significantly associated with breast cancer risk.
CONCLUSION
Our findings indicated that the SNP rs2981582 and haplotypes CGTC, TGTC, and CATC in FGFR2 may be associated with an increased risk of breast cancer in Han Chinese women.
MeSH Terms
-
Asian Continental Ancestry Group*
Breast
Breast Neoplasms
Case-Control Studies*
Female
Fibroblast Growth Factors*
Fibroblasts*
Genetic Variation
Haplotypes
Humans
Polymorphism, Single Nucleotide
Progesterone
Receptor, Fibroblast Growth Factor, Type 2*
Receptors, Fibroblast Growth Factor*
Risk Factors
Fibroblast Growth Factors
Progesterone
Receptor, Fibroblast Growth Factor, Type 2
Receptors, Fibroblast Growth Factor
Figure
Reference
-
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61:69–90.
Article2. Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007; 447:1087–1093.3. Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007; 39:870–874.
Article4. Heath SC, Gut IG, Brennan P, McKay JD, Bencko V, Fabianova E, et al. Investigation of the fine structure of European populations with applications to disease association studies. Eur J Hum Genet. 2008; 16:1413–1429.
Article5. Raskin L, Pinchev M, Arad C, Lejbkowicz F, Tamir A, Rennert HS, et al. FGFR2 is a breast cancer susceptibility gene in Jewish and Arab Israeli populations. Cancer Epidemiol Biomarkers Prev. 2008; 17:1060–1065.
Article6. Lin CY, Ho CM, Bau DT, Yang SF, Liu SH, Lin PH, et al. Evaluation of breast cancer susceptibility loci on 2q35, 3p24, 17q23 and FGFR2 genes in Taiwanese women with breast cancer. Anticancer Res. 2012; 32:475–482.7. Rebbeck TR, DeMichele A, Tran TV, Panossian S, Bunin GR, Troxel AB, et al. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis. 2009; 30:269–274.
Article8. Barnholtz-Sloan JS, Shetty PB, Guan X, Nyante SJ, Luo J, Brennan DJ, et al. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010; 31:1417–1423.
Article9. Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 2005; 16:179–186.
Article10. Moffa AB, Ethier SP. Differential signal transduction of alternatively spliced FGFR2 variants expressed in human mammary epithelial cells. J Cell Physiol. 2007; 210:720–731.
Article11. Moffa AB, Tannheimer SL, Ethier SP. Transforming potential of alternatively spliced variants of fibroblast growth factor receptor 2 in human mammary epithelial cells. Mol Cancer Res. 2004; 2:643–652.12. Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005; 16:139–149.
Article13. Jang JH, Shin KH, Park JG. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res. 2001; 61:3541–3543.14. Johnson BD, Merz CN, Braunstein GD, Berga SL, Bittner V, Hodgson TK, et al. Determination of menopausal status in women: the NHLBI-sponsored Women’s Ischemia Syndrome Evaluation (WISE) Study. J Womens Health (Larchmt). 2004; 13:872–887.
Article15. Liang J, Chen P, Hu Z, Zhou X, Chen L, Li M, et al. Genetic variants in fibroblast growth factor receptor 2 (FGFR2) contribute to susceptibility of breast cancer in Chinese women. Carcinogenesis. 2008; 29:2341–2346.
Article16. Meyer KB, Maia AT, O’Reilly M, Teschendorff AE, Chin SF, Caldas C, et al. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008; 6:e108.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Vitamin D receptor gene polymorphisms in breast cancer
- A case of Pfeiffer syndrome with c833_834GC>TG (Cys278Leu) mutation in the FGFR2 gene
- Association Study of Fibroblast Growth Factor 2 and Fibroblast Growth Factor Receptors Gene Polymorphism in Korean Ossification of the Posterior Longitudinal Ligament Patients
- A case of Apert's Syndrome(Acrocophalosyndactyly) with Fibroblast Growth Factor Receptor 2 Exon IIIa Mutation
- Mutation Analysis in Fibroblast Growth Factor Receptor 3 Gene in Korean Children with Simple Craniosynostosis