J Korean Med Sci.
2006 Oct;21(5):794-799. 10.3346/jkms.2006.21.5.794.
Identification of a Novel Single Nucleotide Polymorphism of HADHA Gene at a Referred Primer-binding Site During Pre-diagnostic Tests for Preimplantation Genetic Diagnosis
- Affiliations
-
- 1Laboratory of Reproductive Biology and Infertility, Cheil General Hospital and Women's Healthcare Center, Sungkyunkwan University School of Medicine, Seoul, Korea. junjh55@hotmail.com
- 2Department of Obstetrics and Gynecology, Cheil General Hospital and Women's Healthcare Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 3Medical Genetics Clinic and Laboratory, Department of Pediatrics, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
- KMID: 1781894
- DOI: http://doi.org/10.3346/jkms.2006.21.5.794
Abstract
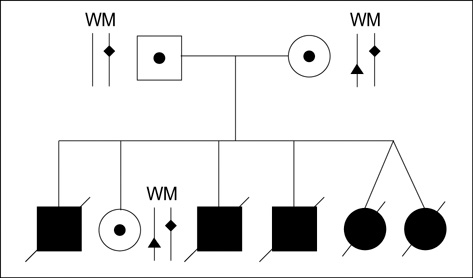
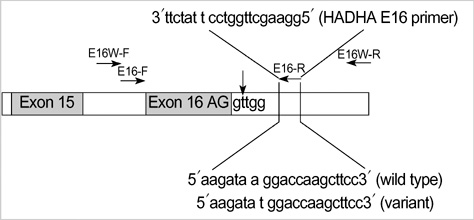
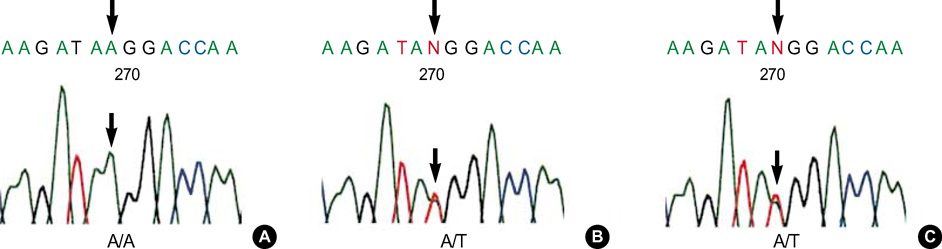
- The pre-diagnostic test for preimplantation genetic diagnosis (PGD) of long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency was performed by polymerase chain reaction (PCR) and direct sequencing for hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (HADHA) gene. We obtained unexpected genotyping results of HADHA gene by allele drop-out in the analysis of patients' genomic DNA samples with a referred PCR primer set. Upon further analysis with a re-designed primer set, we found a novel single nucleotide polymorphism (SNP) at the referred primer-binding site in the normal allele of HADHA gene (NT_022184, 5233296 a>t). We found that the frequency of this novel SNP was 0.064 in Korean population. Pre-diagnostic test using single lymphocytes and clinical PGD were successfully performed with the re-designed primer set. Nineteen embryos (95.0%) among 20 were successfully diagnosed to 5 homozygous mutated, 8 heterozygous carrier and 6 wild type. Among 6 normal embryos, well developed and selected 4 embryos were transferred into the mother's uterus, but a pregnancy was not achieved. We proposed that an unknown SNP at primer-binding sites would be a major cause of allele drop-out in the PGD for single gene dis-order.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Preimplantation Genetic Diagnosis for Ornithine Transcarbamylase Deficiency by Simultaneous Analysis of Duplex-nested PCR and Fluorescence In Situ Hybridization : A Case Report
Hyoung-Song Lee, Jin Hyun Jun, Hye Won Choi, Chun Kyu Lim, Han-Wook Yoo, Mi Kyoung Koong, Inn Soo Kang
J Korean Med Sci. 2007;22(3):572-576. doi: 10.3346/jkms.2007.22.3.572.
Reference
-
1. Lim CK, Jun JH, Min DM, Lee HS, Kim JY, Koong MK, Kang IS. Efficacy and clinical outcome of preimplantation genetic diagnosis using FISH for couples of reciprocal and Robertsonian translocations: the Korean experience. Prenat Diagn. 2004. 24:556–561.2. Lee HS, Choi HW, Lim CK, Min DM, Byun HK, Kim JY, Koong MK, Yoo HW, Kim SC, Jun JH, Kang IS. Successful preimplantation genetic diagnosis for ornithine transcarbamylase deficiency, junctional epidermolysis bullosa and lactic acidosis using duplex nested PCR: delivery of healthy baby by specific preimplantation genetic diagnosis for ornithine transcarbamylase deficiency. Korean J Obstet Gynecol. 2004. 47:708–718.3. Sermon K. Current concepts in preimplantation genetic diagnosis (PGD): a molecular biologist's view. Hum Reprod Update. 2002. 8:11–20.
Article4. Piyamongkol W, Bermudez MG, Harper JC, Wells D. Detailed investigation of factors influencing amplification efficiency and allele drop-out in single cell PCR: implications for preimplantation genetic diagnosis. Mol Hum Reprod. 2003. 9:411–420.
Article5. Findlay I, Ray P, Quirke P, Rutherford A, Lilford R. Allelic drop-out and preferential amplification in single cells and human blastomeres: implications for preimplantation diagnosis of sex and cystic fibrosis. Hum Reprod. 1995. 10:1609–1618.
Article6. Ray PF, Handyside AH. Increasing the denaturation temperature during the first cycles of amplification reduces allele dropout from single cells for preimplantation genetic diagnosis. Mol Hum Reprod. 1996. 2:213–218.7. Thornhill AR, McGrath JA, Eady RA, Braude PR, Handyside AH. A comparison of different lysis buffers to assess allele dropout from single cells for preimplantation genetic diagnosis. Prenat Diagn. 2001. 21:490–497.
Article8. Choi HW, Lee HS, Lim CK, Koong MK, Kang IS, Jun JH. Reliability of the single cell PCR analysis for preimplantation genetic diagnosis of single gene disorders. Korean J Fertil Steril. 2005. 32:293–300.9. Dreesen J, Jacobs L, Bras M, Herberg J, Dumoulin JC, Geraedts JP, Evers JL, Smeets HJ. Multiplex PCR of polymorphic markers flanking the CFTR gene: a general approach for preimplantation genetic diagnosis of cystic fibrosis. Mol Hum Reprod. 2003. 6:391–396.
Article10. Lee HS, Choi HW, Lim CK, Park SY, Kim JY, Koong MK, Jun JH, Kang IS. Efficacy of duplex-nested PCR and fluorescent PCR in the preimplantation genetic diagnosis for Duchenne muscular dystrophy. Korean J Fertil Steril. 2005. 32:17–26.11. Strauss AW, Bennett MJ, Rinaldo P, Sims HF, O'Brien LK, Gibson B, Ibdah J. Inherited long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency and a fetal-maternal interaction cause maternal liver disease and other pregnancy complications. Semin Perinatol. 1999. 23:100–112.
Article12. Gillingham MB, Connor WE, Matern D, Rinaldo P, Burlingame T, Meeuws K, Harding CO. Optimal dietary theraphy of long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency. Mol Genet Metab. 2003. 79:114–123.13. Wong LJ, Chen TJ, Dai P, Bird L, Muenke M. Novel SNP at the common primer site of exon IIIa of FGFR2 gene causes error in molecular diagnosis of Craniosynostosis syndrome. Am J Med Genet. 2001. 102:282–285.14. Solano AR, Dourisboure RJ, Weitzel J, Podesta EJ. A cautionary note: false homozyosity for BRCA2 6174delT mutation resulting from a single nucleotide polymorphism masking the wt allele. Eur J Hum Genet. 2002. 10:395–397.15. Zajickova K, Krepelova A, Zofkova I. A single nucleotide polymorphism under the reverse primer binding site may lead to BsmI misgenotyping in the vitamin D receptor gene. J Bone Miner Res. 2003. 18:1754–1757.
Article16. Leibelt C, Budowle B, Collins P, Daouid Y, Moretti T, Nunn G, Reeder D, Roby R. Identification of a D8S1179 primer binding site mutation and the validation of a primer designed to recover null alleles. Forensic Sci Int. 2003. 133:220–227.
Article17. Verlinsky Y, Rechitsky S, Verlinsky O, Strom C, Kuliev A. Preimplantation diagnosis for long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency. Reprod Biomed Online. 2001. 2:17–19.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Advantages of the single nucleotide polymorphism-based noninvasive prenatal test
- Diagnostic approach for genetic causes of intellectual disability
- Application of Single Nucleotide Polymorphism in Forensic Science
- Genetic association study of single nucleotide polymorphism in dentistry
- An update of preimplantation genetic diagnosis in gene diseases, chromosomal translocation, and aneuploidy screening