Ann Lab Med.
2013 Jan;33(1):14-27. 10.3343/alm.2013.33.1.14.
Rapid Clinical Bacteriology and Its Future Impact
- Affiliations
-
- 1BioMerieux SA, Unit Microbiology, R&D Microbiology, La Balme Les Grottes, France. alex.vanbelkum@biomerieux.com
- 2Geneva University Hospitals, Laboratory of Bacteriology and Genomic Research Laboratory, Geneva, Switzerland.
- 3BioMerieux Inc., Unit Microbiology, Franchise ID/AST, St. Louis, MO, USA.
- KMID: 1781293
- DOI: http://doi.org/10.3343/alm.2013.33.1.14
Abstract
- Clinical microbiology has always been a slowly evolving and conservative science. The sub-field of bacteriology has been and still is dominated for over a century by culture-based technologies. The integration of serological and molecular methodologies during the seventies and eighties of the previous century took place relatively slowly and in a cumbersome fashion. When nucleic acid amplification technologies became available in the early nineties, the predicted "revolution" was again slow but in the end a real paradigm shift did take place. Several of the culture-based technologies were successfully replaced by tests aimed at nucleic acid detection. More recently a second revolution occurred. Mass spectrometry was introduced and broadly accepted as a new diagnostic gold standard for microbial species identification. Apparently, the diagnostic landscape is changing, albeit slowly, and the combination of newly identified infectious etiologies and the availability of innovative technologies has now opened new avenues for modernizing clinical microbiology. However, the improvement of microbial antibiotic susceptibility testing is still lagging behind. In this review we aim to sketch the most recent developments in laboratory-based clinical bacteriology and to provide an overview of emerging novel diagnostic approaches.
Keyword
MeSH Terms
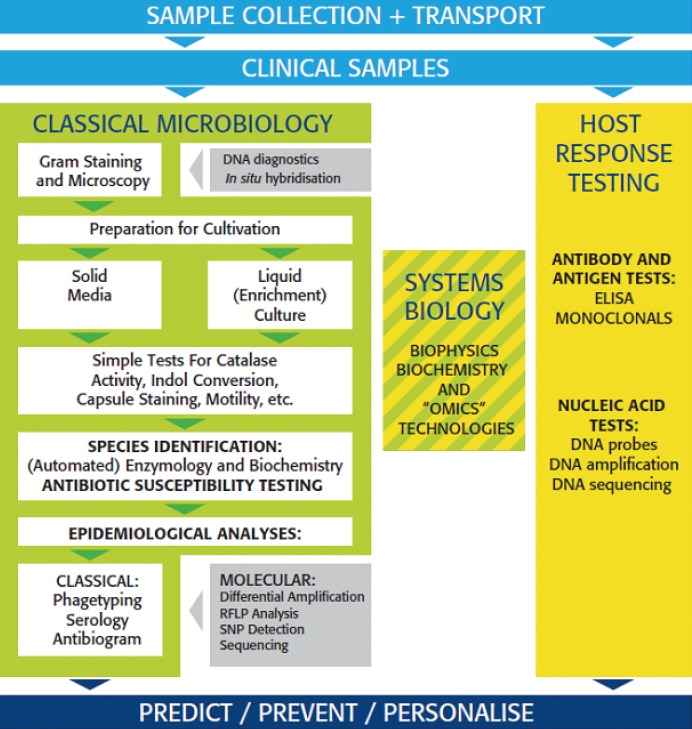
Figure
Reference
-
1. O'hara CM. Manual and automated instrumentation for identification of Enterobacteriaceae and other aerobic gram-negative bacilli. Clin Microbiol Rev. 2005; 18:147–162. PMID: 15653824.2. Treviño M, Areses P, Peñalver MD, Cortizo S, Pardo F, del Molino ML, et al. Susceptibility trends of Bacteroides fragilis group and characterisation of carbapenemase-producing strains by automated REP-PCR and MALDI-TOF. Anaerobe. 2012; 18:37–43. PMID: 22261518.3. Saravolatz LD, Pawlak J, Johnson LB. In Vitro susceptibilities and molecular analysis of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus isolates. Clin Infect Dis. 2012; 55:582–586. PMID: 22615331.4. Steingart KR, Henry M, Laal S, Hopewell PC, Ramsay A, Menzies D, et al. Commercial serological antibody detection tests for the diagnosis of pulmonary tuberculosis: a systematic review. PLoS Med. 2007; 4:e202. PMID: 17564490.
Article5. Wattiau P, Boland C, Bertrand S. Methodologies for Salmonella enterica subsp. enterica subtyping: gold standards and alternatives. Appl Environ Microbiol. 2011; 77:7877–7885. PMID: 21856826.6. Fredlund H, Falk L, Jurstrand M, Unemo M. Molecular genetic methods for diagnosis and characterisation of Chlamydia trachomatis and Neisseria gonorrhoeae: impact on epidemiological surveillance and interventions. APMIS. 2004; 112:771–784. PMID: 15638837.7. Hadgu A, Dendukuri N, Hilden J. Evaluation of nucleic acid amplification tests in the absence of a perfect gold-standard test: a review of the statistical and epidemiologic issues. Epidemiology. 2005; 16:604–612. PMID: 16135935.8. Van Belkum A, Welker M, Erhard M, Chatellier S. Biomedical mass spectrometry in today's and tomorrow's clinical microbiology laboratories. J Clin Microbiol. 2012; 50:1513–1517. PMID: 22357505.
Article9. Zhang W, Li F, Nie L. Integrating multiple 'omics' analysis for microbial biology: application and methodologies. Microbiology. 2010; 156:287–301. PMID: 19910409.
Article10. Kerremans JJ, Verbrugh HA, Vos MC. Frequency of microbiologically correct antibiotic therapy increased by infectious disease consultations and microbiological results. J Clin Microbiol. 2012; 50:2066–2068. PMID: 22422850.
Article11. Galar A, Keiva J, Espinosa M, Guillen-Grima F, Hernaez S, Yuste JR. Clinical and economic evaluation of the impact of rapid microbiological diagnostic testing. J Infect. 2012; 6. 19. [Epub ahead of print].
Article12. Kerremans JJ, van der Bij AK, Goessens W, Verbrugh HA, Vos MC. Needle-to-incubator transport time: logistic factors influencing transport time for blood culture specimens. J Clin Microbiol. 2009; 47:819–822. PMID: 19129412.
Article13. Griffiths LJ, Anyim M, Doffman SR, Wilks M, Millar MR, Agrawal SG. Comparison of DNA extraction methods for Aspergillus fumigatus using real-time PCR. J Med Microbiol. 2006; 55:1187–1191. PMID: 16914647.14. Dauphin LA, Walker RE, Petersen JM, Bowen MD. Comparative evaluation of automated and manual commercial DNA extraction methods for detection of Francisella tularensis DNA from suspensions and spiked swabs by real-time polymerase chain reaction. Diagn Microbiol Infect Dis. 2011; 70:299–306. PMID: 21546201.15. Barczak AK, Gomez JE, Kaufmann BB, Hinson ER, Cosimi L, Borowsky ML, et al. RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities. Proc Natl Acad Sci U S A. 2012; 109:6217–6222. PMID: 22474362.
Article16. Ingham C, Schneeberger . Hays , Van Leeuwen , editors. Can we improve on the Petri dish with porous culture support? Bentham Science Publisher eBook series. 2012. p. 3–16.17. Ingham CJ, Boonstra S, Levels S, de Lange M, Meis JF, Schneeberger PM. Rapid susceptibility testing and microcolony analysis of Candida spp. cultured and imaged on porous aluminum oxide. PLoS One. 2012; 7:e33818. PMID: 22439000.18. Váradi L, Gray M, Groundwater PW, Hall AJ, James AL, Orenga S, et al. Synthesis and evaluation of fluorogenic 2-amino-1,8-naphthyridine derivatives for the detection of bacteria. Org Biomol Chem. 2012; 10:2578–2589. PMID: 22354016.
Article19. Van Hoecke F, Deloof N, Claeys G. Performance evaluation of a modified chromogenic medium, ChromID MRSA New, for the detection of methicillin-resistant Staphylococcus aureus from clinical specimens. Eur J Clin Microbiol Infect Dis. 2011; 30:1595–1598. PMID: 21499707.20. Nordmann P, Girlich D, Poirel L. Detection of carbapenemase producers in enterobacteriaceae by use of a novel screening medium. J Clin Microbiol. 2012; 50:2761–2766. PMID: 22357501.
Article21. Garrec H, Drieux-Rouzet L, Golmard JL, Jarlier V, Robert J. Comparison of nine phenotypic methods for detection of extended-spectrum beta-lactamase production by Enterobacteriaceae. J Clin Microbiol. 2011; 49:1048–1057. PMID: 21248086.22. London R, Schwedock J, Sage A, Valley H, Meadows J, Waddington M, et al. An automated system for rapid non-destructive enumeration of growing microbes. PLoS One. 2010; 5:e8609. PMID: 20062794.
Article23. Towns ML, Jarvis WR, Hsueh PR. Guidelines on blood cultures. J Microbiol Immunol Infect. 2010; 43:347–349. PMID: 20688297.
Article24. Jin WY, Jang SJ, Lee MJ, Park G, Kim MJ, Kook JK, et al. Evaluation of VITEK2, MicroScan, and Phoenix for identification of clinical isolates and reference strains. Diagn Microbiol Infect Dis. 2011; 70:442–447. PMID: 21767700.25. Doern GV, Scott DR, Rashad AL. Clinical impact of rapid antimicrobial susceptibility testing of blood culture isolates. Antimicrob Agents Chemother. 1982; 21:1023–1024. PMID: 7114834.
Article26. Kerremans JJ, Verboom P, Stijnen T, Hakkaart-van Roijen L, Goessens W, Verbrugh HA, et al. Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J Antimicrob Chemother. 2008; 61:428–435. PMID: 18156278.
Article27. Horn JE, Quinn T, Hammer M, Palmer L, Falkow S. Use of nucleic acid probes for the detection of sexually transmitted infectious agents. Diagn Microbiol Infect Dis. 1986; 4(3 Suppl):101S–109S. PMID: 3084160.
Article28. Mulks MH, Simpson DA, Shoberg RJ. Restriction site polymorphism in genes encoding type 2 but not type 1 gonococcal IgA1 proteases. Antonie Van Leeuwenhoek. 1987; 53:471–478. PMID: 2897189.
Article29. Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, et al. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003; 41:1574–1585. PMID: 12682148.30. Rank EL, Sautter RL, Beavis KG, Harris S, Jones S, Drechsel R, et al. A two-site analytical evaluation of the BD Viper System with XTR technology in non-extracted mode and extracted mode with seeded simulated specimens. J Lab Autom. 2011; 16:271–275. PMID: 21764022.
Article31. Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010; 363:1005–1015. PMID: 20825313.
Article32. Ecker DJ, Sampath R, Li H, Massire C, Matthews HE, Toleno D, et al. New technology for rapid molecular diagnosis of bloodstream infections. Expert Rev Mol Diagn. 2010; 10:399–415. PMID: 20465496.
Article33. Tenover FC. Developing molecular amplification methods for rapid diagnosis of respiratory tract infections caused by bacterial pathogens. Clin Infect Dis. 2011; 52(Suppl 4):S338–S345. PMID: 21460293.
Article34. Emmadi R, Boonyaratanakornkit JB, Selvarangan R, Shyamala V, Zimmer BL, Williams L, et al. Molecular methods and platforms for infectious diseases testing a review of FDA-approved and cleared assays. J Mol Diagn. 2011; 13:583–604. PMID: 21871973.35. van Hal SJ, Jennings Z, Stark D, Marriott D, Harkness J. MRSA detection: comparison of two molecular methods (BD GeneOhm PCR assay and Easy-Plex) with two selective MRSA agars (MRSA-ID and Oxoid MRSA) for nasal swabs. Eur J Clin Microbiol Infect Dis. 2009; 28:47–53. PMID: 18663499.
Article36. Van Belkum A, Melles DC, Nouwen J, van Leeuwen WB, van Wamel W, Vos MC, et al. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect Genet Evol. 2009; 9:32–47. PMID: 19000784.37. Angione SL, Chauhan A, Tripathi A. Real-time droplet DNA amplification with a new tablet platform. Anal Chem. 2012; 84:2654–2661. PMID: 22320164.
Article38. Hatch AC, Fisher JS, Tovar AR, Hsieh AT, Lin R, Pentoney SL, et al. 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab Chip. 2011; 11:3838–3845. PMID: 21959960.
Article39. DeGrasse JA. A single-stranded DNA aptamer that selectively binds to Staphylococcus aureus enterotoxin B. PLoS One. 2012; 7:e33410. PMID: 22438927.40. Syrmis MW, Moser RJ, Whiley DM, Vaska V, Coombs GW, Nissen MD, et al. Comparison of a multiplexed MassARRAY system with real-time allele-specific PCR technology for genotyping of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2011; 17:1804–1810. PMID: 21595795.41. Ivanov PL. A new approach to forensic medical typing of human mitochondrial DNA with the use of mass-spectrometric analysis of amplified fragments: PLEX-ID automated genetic analysis system. Sud Med Ekspert. 2010; 53:46–51. PMID: 20734792.42. Shitikov E, Ilina E, Chernousova L, Borovskaya A, Rukin I, Afanas'ev M, et al. Mass spectrometry based methods for the discrimination and typing of mycobacteria. Infect Genet Evol. 2012; 12:838–845. PMID: 22230718.
Article43. Hrabák J, Walková R, Studentová V, Chudácková E, Bergerová T. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011; 49:3222–3227. PMID: 21775535.
Article44. Burckhardt I, Zimmermann S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J Clin Microbiol. 2011; 49:3321–3324. PMID: 21795515.
Article45. Hooff GP, van Kampen JJ, Meesters RJ, van Belkum A, Goessens WH, Luider TM. Characterization of β-lactamase enzyme activity in bacterial lysates using MALDI-mass spectrometry. J Proteome Res. 2012; 11:79–84. PMID: 22013912.
Article46. Hrabák J, Studentová V, Walková R, Zemlicková H, Jakubu V, Chudácková E, et al. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 Carbapenemases by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J Clin Microbiol. 2012; 50:2441–2443. PMID: 22553235.47. Marinach C, Alanio A, Palous M, Kwasek S, Fekkar A, Brossas JY, et al. MALDI-TOF MS-based drug susceptibility testing of pathogens: the example of Candida albicans and fluconazole. Proteomics. 2009; 9:4627–4627. PMID: 19750514.48. De Carolis E, Vella A, Florio AR, Posteraro P, Perlin DS, Sanguinetti M, et al. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. J Clin Microbiol. 2012; 50:2479–2483. PMID: 22535984.49. Kaman WE, Galassi F, de Soet JJ, Bizzarro S, Loos BG, Veerman EC, et al. Highly specific protease-based approach for detection of Porphyromonas gingivalis in diagnosis of periodontitis. J Clin Microbiol. 2012; 50:104–112. PMID: 22075590.50. Kaman WE, Hulst AG, van Alphen PT, Roffel S, van der, Merkel T, et al. Peptide-based fluorescence resonance energy transfer protease substrates for the detection and diagnosis of Bacillus species. Anal Chem. 2011; 83:2511–2517. PMID: 21370823.51. Köhling HL, Bittner A, Müller KD, Buer J, Becker M, Rübben H, et al. Direct identification of bacteria in urine samples by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and relevance of defensins as interfering factors. J Med Microbiol. 2012; 61:339–344. PMID: 22275503.
Article52. Everley RA, Mott TM, Wyatt SA, Toney DM, Croley TR. Liquid chromatography/mass spectrometry characterization of Escherichia coli and Shigella species. J Am Soc Mass Spectrom. 2008; 19:1621–1628. PMID: 18692404.53. Turner KM. The secret life of the multilocus sequence type. Int J Antimicrob Agents. 2007; 29:129–135. PMID: 17204401.
Article54. Relman DA. Microbial genomics and infectious diseases. N Engl J Med. 2011; 365:347–357. PMID: 21793746.
Article55. Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995; 269:496–512. PMID: 7542800.56. Eyre DW, Golubchik T, Gordon NC, Bowden R, Piazza P, Batty EM, et al. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open. 2012; 2:e001124.57. Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med. 2012; 366:2267–2275. PMID: 22693998.
Article58. Lam HY, Clark MJ, Chen R, Chen R, Natsoulis G, O'Huallachain M, et al. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol. 2011; 30:78–82. PMID: 22178993.
Article59. Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2011; 13:47–58. PMID: 22179717.
Article60. Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008; 24:4–10. PMID: 18043225.
Article61. Sommer MO, Dantas G. Antibiotics and the resistant microbiome. Curr Opin Microbiol. 2011; 14:556–563. PMID: 21802347.
Article62. Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012; 30:434–439. PMID: 22522955.
Article63. Korpi A, Järnberg J, Pasanen AL. Microbial volatile organic compounds. Crit Rev Toxicol. 2009; 39:139–193. PMID: 19204852.
Article64. Mgode GF, Weetjens BJ, Nawrath T, Cox C, Jubitana M, Machang'u RS, et al. Diagnosis of tuberculosis by trained African giant pouched rats and confounding impact of pathogens and microflora of the respiratory tract. J Clin Microbiol. 2012; 50:274–280. PMID: 22135255.
Article65. Bruins M, Bos A, Petit PL, Eadie K, Rog A, Bos R, et al. Device-independent, real-time identification of bacterial pathogens with a metal oxide-based olfactory sensor. Eur J Clin Microbiol Infect Dis. 2009; 28:775–780. PMID: 19190942.
Article66. McNerney R, Mallard K, Okolo PI, Turner C. Production of volatile organic compounds by mycobacteria. FEMS Microbiol Lett. 2012; 328:150–156. PMID: 22224870.
Article67. Crespo E, Cristescu SM, de Ronde H, Kuijper S, Kolk AH, Anthony RM, et al. Proton Transfer Reaction Mass Spectrometry detects rapid changes in volatile metabolite emission by Mycobacterium smegmatis after the addition of specific antimicrobial agents. J Microbiol Methods. 2011; 86:8–15. PMID: 21277343.68. Beekes M, Lasch P, Naumann D. Analytical applications of Fourier transform-infrared (FT-IR) spectroscopy in microbiology and prion research. Vet Microbiol. 2007; 123:305–319. PMID: 17540519.
Article69. Maquelin K, Kirchner C, Choo-Smith LP, van den Braak N, Endtz HP, Naumann D, et al. Identification of medically relevant microorganisms by vibrational spectroscopy. J Microbiol Methods. 2002; 51:255–271. PMID: 12223286.
Article70. Shachaf CM, Elchuri SV, Koh AL, Zhu J, Nguyen LN, Mitchell DJ, et al. A novel method for detection of phosphorylation in single cells by surface enhanced Raman scattering (SERS) using composite organic-inorganic nanoparticles (COINs). PLoS One. 2009; 4:e5206. PMID: 19367337.
Article71. Orsini F, Ami D, Villa AM, Sala G, Bellotti MG, Doglia SM. FT-IR microspectroscopy for microbiological studies. J Microbiol Methods. 2000; 42:17–27. PMID: 11000427.
Article72. Willemse-Erix D, Bakker-Schut T, Slagboom-Bax F, Jachtenberg JW, Lemmens-den Toom N, Papagiannitsis CC, et al. Rapid typing of extended-spectrum β-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolates by use of SpectraCell RA. J Clin Microbiol. 2012; 50:1370–1375. PMID: 22238437.73. Wulf MW, Willemse-Erix D, Verduin CM, Puppels G, van Belkum A, Maquelin K. The use of Raman spectroscopy in the epidemiology of methicillin-resistant Staphylococcus aureus of human- and animal-related clonal lineages. Clin Microbiol Infect. 2012; 18:147–152. PMID: 21854500.74. Willemse-Erix DF, Jachtenberg JW, Schut TB, van Leeuwen W, van Belkum A, Puppels G, et al. Towards Raman-based epidemiological typing of Pseudomonas aeruginosa. J Biophotonics. 2010; 3:506–511. PMID: 20572285.75. Willemse-Erix HF, Jachtenberg J, Barutçi H, Puppels GJ, van Belkum A, Vos MC, et al. Proof of principle for successful characterization of methicillin-resistant coagulase-negative staphylococci isolated from skin by use of Raman spectroscopy and pulsed-field gel electrophoresis. J Clin Microbiol. 2010; 48:736–740. PMID: 20042618.
Article76. Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med. 2011; 17:1015–1019. PMID: 21804541.
Article77. Joon Tam Y, Mohd Lila MA, Bahaman AR. Development of solid - based paper strips for rapid diagnosis of Pseudorabies infection. Trop Biomed. 2004; 21:121–134. PMID: 16493404.78. Thornton CR. Development of an immunochromatographic lateral-flow device for rapid serodiagnosis of invasive aspergillosis. Clin Vaccine Immunol. 2008; 15:1095–1105. PMID: 18463222.
Article79. Lafleur L, Stevens D, McKenzie K, Ramachandran S, Spicar-Mihalic P, Singhal M, et al. Progress toward multiplexed sample-to-result detection in low resource settings using microfluidic immunoassay cards. Lab Chip. 2012; 12:1119–1127. PMID: 22311085.
Article80. Shinde SB, Fernandes CB, Patravale VB. Recent trends in in-vitro nanodiagnostics for detection of pathogens. J Control Release. 2011; 159:164–180. PMID: 22192572.
Article81. Siddiqui S, Dai Z, Stavis CJ, Zeng H, Moldovan N, Hamers RJ, et al. A quantitative study of detection mechanism of a label-free impedance biosensor using ultrananocrystalline diamond microelectrode array. Biosens Bioelectron. 2012; 35:284–290. PMID: 22456097.
Article82. Chang TC, Huang AH. Rapid differentiation of fermentative from non-fermentative gram-negative bacilli in positive blood cultures by an impedance method. J Clin Microbiol. 2000; 38:3589–3594. PMID: 11015369.
Article83. Wu JJ, Huang AH, Dai JH, Chang TC. Rapid detection of oxacillin-resistant Staphylococcus aureus in blood cultures by an impedance method. J Clin Microbiol. 1997; 35:1460–1464. PMID: 9163462.84. Walsh TR, Toleman MA. The emergence of pan-resistant Gram-negative pathogens merits a rapid global political response. J Antimicrob Chemother. 2012; 67:1–3. PMID: 21994911.
Article85. Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010; 74:417–433. PMID: 20805405.
Article86. Lee HH, Collins JJ. Microbial environments confound antibiotic efficacy. Nat Chem Biol. 2011; 8:6–9. PMID: 22173343.
Article87. Belenky P, Collins JJ. Microbiology. Antioxidant strategies to tolerate antibiotics. Science. 2011; 334:915–916. PMID: 22096180.88. Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011; 473:216–220. PMID: 21562562.
Article89. Waldeisen JR, Wang T, Mitra D, Lee LP. A real-time PCR antibiogram for drug-resistant sepsis. PLoS One. 2011; 6:e28528. PMID: 22164303.
Article90. Beuving J, Verbon A, Gronthoud FA, Stobberingh EE, Wolffs PF. Antibiotic susceptibility testing of grown blood cultures by combining culture and real-time polymerase chain reaction is rapid and effective. PLoS One. 2011; 6:e27689. PMID: 22194790.
Article91. Jorgensen JH, Ferraro MJ. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis. 2009; 49:1749–1755. PMID: 19857164.
Article92. Douglas IS, Price C, Overdier K, Thompson K, Wolken B, Metzger S, et al. Rapid Microbiological Identification and Major Drug Resistance Phenotyping with Novel Multiplexed Automated Digital Microscopy (MADM) for Ventilator-associated Pneumonia (VAP) Surveillance. 2011. Updated in 2012. In : American Thoracic Society; Denver, CO.. http://www.accelr8.com/docs/ATS_2011a.pdf.93. Cronin UP, Wilkinson MG. The potential of flow cytometry in the study of Bacillus cereus. J Appl Microbiol. 2010; 108:1–16. PMID: 19486207.94. Filoche SK, Coleman MJ, Angker L, Sissons CH. A fluorescence assay to determine the viable biomass of microcosm dental plaque biofilms. J Microbiol Methods. 2007; 69:489–496. PMID: 17408789.
Article95. Shrestha NK, Scalera NM, Wilson DA, Procop GW. Rapid differentiation of methicillin-resistant and methicillin-susceptible Staphylococcus aureus by flow cytometry after brief antibiotic exposure. J Clin Microbiol. 2011; 49:2116–2120. PMID: 21471343.96. Beech JP, Holm SH, Adolfsson K, Tegenfeldt JO. Sorting cells by size, shape and deformability. Lab Chip. 2012; 12:1048–1051. PMID: 22327631.
Article97. Godin M, Delgado FF, Son S, Grover WH, Bryan AK, Tzur A, et al. Using buoyant mass to measure the growth of single cells. Nat Methods. 2010; 7:387–390. PMID: 20383132.
Article98. Knudsen SM, von Muhlen MG, Schauer DB, Manalis SR. Determination of bacterial antibiotic resistance based on osmotic shock response. Anal Chem. 2009; 81:7087–7090. PMID: 20337387.
Article99. Howell M, Wirz D, Daniels AU, Braissant O. Application of a micro-calorimetric method for determining drug susceptibility in Mycobacterium species. J Clin Microbiol. 2012; 50:16–20. PMID: 22090404.100. Bonkat G, Braissant O, Widmer AF, Frei R, Rieken M, Wyler S, et al. Rapid detection of urinary tract pathogens using micro-calorimetry principle, technique and first results. BJU Int. 2012; 2. 07. [Epub ahead of print].
Article101. Buchholz F, Wolf A, Lerchner J, Mertens F, Harms H, Maskow T. Chip calorimetry for fast and reliable evaluation of bactericidal and bacteriostatic treatments of biofilms. Antimicrob Agents Chemother. 2010; 54:312–319. PMID: 19822705.
Article102. Ohlinger A, Deak A, Lutich AA, Feldmann J. Optically trapped gold nanoparticle enables listening at the microscale. Phys Rev Lett. 2012; 108:018101. PMID: 22304294.
Article103. Kaittanis C, Boukhriss H, Santra S, Naser SA, Perez JM. Rapid and sensitive detection of an intracellular pathogen in human peripheral leukocytes with hybridizing magnetic relaxation nanosensors. PLoS One. 2012; 7:e35326. PMID: 22496916.
Article104. Kinnunen P, McNaughton BH, Albertson T, Sinn I, Mofakham S, Elbez R, et al. Self-assembled magnetic bead biosensor for measuring bacterial growth and antimicrobial susceptibility testing. Small. 2012; 6. 05. [Epub ahead of print].
Article105. Fischer A, Yang SJ, Bayer AS, Vaezzadeh AR, Herzig S, Stenz L, et al. Daptomycin resistance mechanisms in clinically derived Staphylococcus aureus strains assessed by a combined transcriptomics and proteomics approach. J Antimicrob Chemother. 2011; 66:1696–1711. PMID: 21622973.106. Sangurdekar DP, Srienc F, Khodursky AB. A classification based framework for quantitative description of large-scale microarray data. Genome Biol. 2006; 7:R32. PMID: 16626502.107. Brazas MD, Hancock RE. Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2005; 49:3222–3227. PMID: 16048929.108. Delmas T, Piraux H, Couffin AC, Texier I, Vinet F, Poulin P, et al. How to prepare and stabilize very small nanoemulsions. Langmuir. 2011; 27:1683–1692. PMID: 21226496.
Article109. Baraban L, Bertholle F, Salverda ML, Bremond N, Panizza P, Baudry J, et al. Millifluidic droplet analyser for microbiology. Lab Chip. 2011; 11:4057–4062. PMID: 22012599.
Article110. Boitard L, Cottinet D, Kleinschmitt C, Bremond N, Baudry J, Yvert G, et al. Monitoring single-cell bioenergetics via the coarsening of emulsion droplets. Proc Natl Acad Sci U S A. 2012; 109:7181–7186. PMID: 22538813.
Article111. Wilson SM, al-Suwaidi Z, McNerney R, Porter J, Drobniewski . Evaluation of a new rapid bacteriophage-based method for the drug susceptibility testing of Mycobacterium tuberculosis. Nat Med. 1997; 3:465–468. PMID: 9095184.112. Eltringham IJ, Wilson SM, Drobniewski FA. Evaluation of a bacteriophage-based assay (phage amplified biologically assay) as a rapid screen for resistance to isoniazid, ethambutol, streptomycin, pyrazinamide, and ciprofloxacin among clinical isolates of Mycobacterium tuberculosis. J Clin Microbiol. 1999; 37:3528–3532. PMID: 10523547.113. Banaiee N, Bobadilla-Del-Valle M, Bardarov S Jr, Riska PF, Small PM, Ponce-De-Leon A, et al. Luciferase reporter mycobacteriophages for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis in Mexico. J Clin Microbiol. 2001; 39:3883–3888. PMID: 11682502.114. Hazbón MH, Guarín N, Ferro BE, Rodríguez AL, Labrada LA, Tovar R, et al. Photographic and luminometric detection of luciferase reporter phages for drug susceptibility testing of clinical Mycobacterium tuberculosis isolates. J Clin Microbiol. 2003; 41:4865–4869. PMID: 14532245.115. Banaiee N, Bobadilla-del-Valle M, Riska PF, Bardarov S Jr, Small PM, Ponce-de-Leon A, et al. Rapid identification and susceptibility testing of Mycobacterium tuberculosis from MGIT cultures with luciferase reporter mycobacteriophages. J Med Microbiol. 2003; 52:557–561. PMID: 12808076.116. Smietana M, Bock WJ, Mikulic P, Ng A, Chinnappan R, Zourob M. Detection of bacteria using bacteriophages as recognition elements immobilized on long-period fiber gratings. Opt Express. 2011; 19:7971–7978. PMID: 21643046.
Article117. Traore H, Ogwang S, Mallard K, Joloba ML, Mumbowa F, Narayan K, et al. Low-cost rapid detection of rifampicin resistant tuberculosis using bacteriophage in Kampala, Uganda. Ann Clin Microbiol Antimicrob. 2007; 6:1. PMID: 17212825.
Article118. Piuri M, Jacobs WR Jr, Hatfull GF. Fluoromycobacteriophages for rapid, specific, and sensitive antibiotic susceptibility testing of Mycobacterium tuberculosis. PLoS One. 2009; 4:e4870. PMID: 19300517.119. Pholwat S, Ehdaie B, Foongladda S, Kelly K, Houpt E. Real-time PCR using mycobacteriophage DNA for rapid phenotypic drug susceptibility results for Mycobacterium tuberculosis. J Clin Microbiol. 2012; 50:754–761. PMID: 22170929.120. Mole R, Trollip A, Abrahams C, Bosman M, Albert H. Improved contamination control for a rapid phage-based rifampicin resistance test for Mycobacterium tuberculosis. J Med Microbiol. 2007; 56:1334–1339. PMID: 17893170.121. Dwyer DJ, Camacho DM, Kohanski MA, Callura JM, Collins JJ. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol Cell. 2012; 46:561–572. PMID: 22633370.
Article122. Cira NJ, Ho JY, Dueck ME, Weibel DB. A self-loading microfluidic device for determining the minimum inhibitory concentration of antibiotics. Lab Chip. 2012; 12:1052–1059. PMID: 22193301.
Article123. Ding X, Lin SC, Kiraly B, Yue H, Li S, Chiang IK, et al. On-chip manipulation of single microparticles, cells, and organisms using surface acoustic waves. Proc Natl Acad Sci U S A. 2012; 109:11105–11109. PMID: 22733731.
Article124. Scholl D, Gebhart D, Williams SR, Bates A, Mandrell R. Genome sequence of E. coli O104:H4 leads to rapid development of a targeted antimicrobial agent against this emerging pathogen. PLoS One. 2012; 7:e33637. PMID: 22432037.
Article125. Contag CH. In vivo pathology: seeing with molecular specificity and cellular resolution in the living body. Annu Rev Pathol. 2007; 2:277–305. PMID: 18039101.
Article126. Rieder R, Zhao Z, Nittayajarn A, Zavizion B. Direct detection of the bacterial stress response in intact samples of platelets by differential impedance. Transfusion. 2011; 51:1037–1046. PMID: 20977486.
Article127. Halouska S, Fenton RJ, Barletta RG, Powers R. Predicting the in vivo mechanism of action for drug leads using NMR metabolomics. ACS Chem Biol. 2012; 7:166–171. PMID: 22007661.
Article128. Greub G, Prod'hom G. Automation in clinical bacteriology: what system to choose? Clin Microbiol Infect. 2011; 17:655–660. PMID: 21521409.
Article129. Matthews S, Deutekom J. The future of diagnostic bacteriology. Clin Microbiol Infect. 2011; 17:651–654. PMID: 21521408.
Article130. Goff DA. iPhones, iPads and medical applications for antimicrobial stewardship. Pharmacotherapy. 2012; 32:657–661. PMID: 22605544.
Article131. Lin Y, Trouillon R, Safina G, Ewing AG. Chemical analysis of single cells. Anal Chem. 2011; 83:4369–4392. PMID: 21500835.
Article132. Mori Y, Miyawaki K, Kato K, Takenaka K, Iwasaki H, Harada N, et al. Diagnostic value of serum procalcitonin and C-reactive protein for infections after allogeneic hematopoietic stem cell transplantation versus non-transplant setting. Intern Med. 2011; 50:2149–2155. PMID: 21963733.
Article133. Fowler CL. Procalcitonin for triage of patients with respiratory tract symptoms: a case study in the trial design process for approval of a new diagnostic test for lower respiratory tract infections. Clin Infect Dis. 2011; 52(Suppl 4):S351–S356. PMID: 21460295.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Revolutionising Bacteriology to Improve Treatment Outcomes and Antibiotic Stewardship
- Bacteriology of the biliary tract
- Microbial Genome Analysis and Application to Clinical Bateriology
- Bacteriology of the Postoperative Mucocele of the Maxillary Sinus
- Message from the Precision and Future Medicine editors to our ad hoc reviewers