Immune Netw.
2009 Apr;9(2):35-40. 10.4110/in.2009.9.2.35.
Rap Signaling in Normal Lymphocyte Development and Leukemia Genesis
- Affiliations
-
- 1Department of Immunology and Cell Biology, Graduate School of Medicine, Kyoto University, Sakyo-ku, Kyoto, Japan. minato@imm.med.kyoto-u.ac.jp
- KMID: 1474592
- DOI: http://doi.org/10.4110/in.2009.9.2.35
Abstract
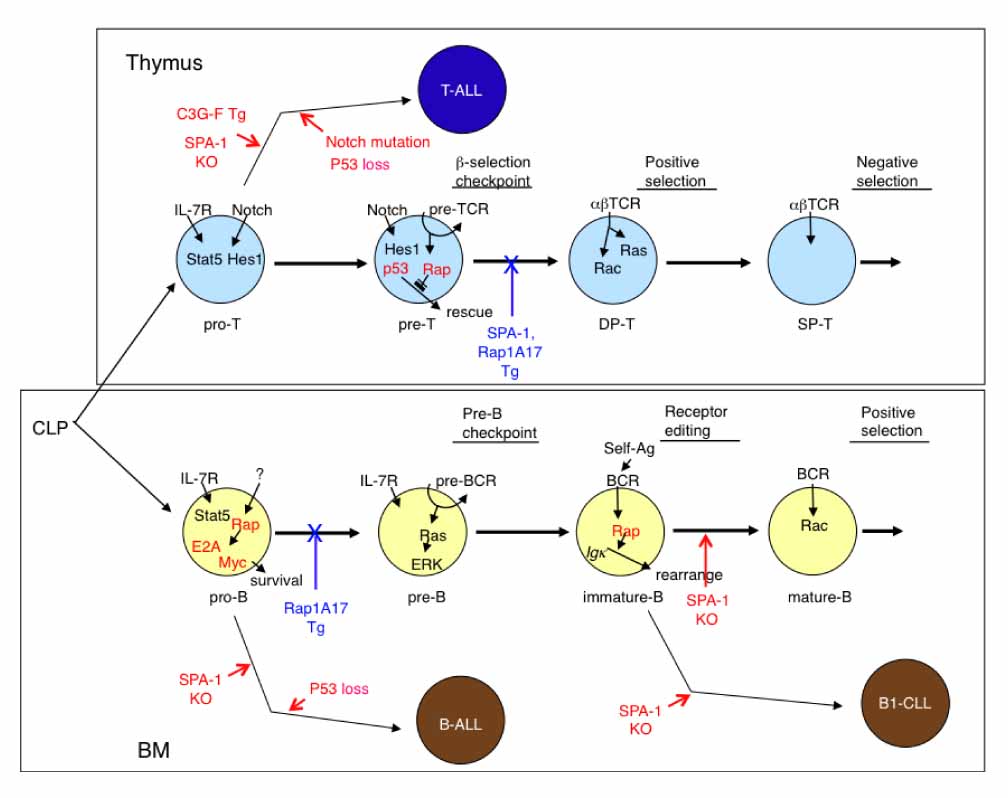
- Although Rap GTPases of the Ras family remained enigmatic for years, extensive studies in this decade have revealed diverse functions of Rap signaling in the control of cell proliferation, differentiation, survival, adhesion, and movement. With the use of gene-engineered mice, we have uncovered essential roles of endogenous Rap signaling in normal lymphocyte development of both T- and B-lineage cells. Deregulation of Rap signaling, on the other hand, results in the development of characteristic leukemia in manners highly dependent on the contexts of cell lineages. These results highlight crucial roles of Rap signaling in the physiology and pathology of lymphocyte development.
Keyword
MeSH Terms
Figure
Reference
-
1. Pizon V, Chardin P, Lerosey I, Olofsson B, Tavitian A. Human cDNAs rap1 and rap2 homologous to the Drosophila gene Dras3 encode proteins closely related to ras in the 'effector' region. Oncogene. 1988. 3:201–214.2. Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol. 2001. 2:369–377.
Article3. Hattori M, Minato N. Rap1 GTPase: functions, regulation, and malignancy. J Biochem. 2003. 134:479–484.
Article4. Sasagawa S, Ozaki Y, Fujita K, Kuroda S. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat Cell Biol. 2005. 7:365–373.
Article5. Minato N, Kometani K, Hattori M. Regulation of immune responses and hematopoiesis by the Rap1 signal. Adv Immunol. 2007. 93:229–264.
Article6. Tsukamoto N, Hattori M, Yang H, Bos JL, Minato N. Rap1 GTPase-activating protein SPA-1 negatively regulates cell adhesion. J Biol Chem. 1999. 274:18463–18469.
Article7. Katagiri K, Hattori M, Minato N, Irie S, Takatsu K, Kinashi T. Rap1 is a potent activation signal for leukocyte function-associated antigen 1 distinct from protein kinase C and phosphatidylinositol-3-OH kinase. Mol Cell Biol. 2000. 20:1956–1969.
Article8. Bos JL. Linking Rap to cell adhesion. Curr Opin Cell Biol. 2005. 17:123–128.
Article9. Katagiri K, Maeda A, Shimonaka M, Kinashi T. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat Immunol. 2003. 4:741–748.
Article10. Duchniewicz M, Zemojtel T, Kolanczyk M, Grossmann S, Scheele JS, Zwartkruis FJ. Rap1A-deficient T and B cells show impaired integrin-mediated cell adhesion. Mol Cell Biol. 2006. 26:643–653.
Article11. Chen Y, Yu M, Podd A, Wen R, Chrzanowska-Wodnicka M, White GC, Wang D. A critical role of Rap1b in B-cell trafficking and marginal zone B-cell development. Blood. 2008. 111:4627–4636.
Article12. Dupuy AG, L'Hoste S, Cherfils J, Camonis J, Gaudriault G, de Gunzburg J. Novel Rap1 dominant-negative mutants interfere selectively with C3G and Epac. Oncogene. 2005. 24:4509–4520.
Article13. Kometani K, Moriyama M, Nakashima Y, Katayama Y, Wang S-F, Saito T, Hattori M, Minato N. Essential role of Rap signal in pre-TCR-mediated β-selection checkpoint in alphabeta T cell development. Blood. 2008. 112:4565–4573.
Article14. Ciofani M, Schmitt TM, Ciofani A, Michie AM, Cuburu N, Aublin A, Maryanski JL, Zúñiga-Pflücker JC. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J Immunol. 2004. 172:5230–5239.
Article15. Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, Pear WS. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med. 2006. 203:2239–2245.
Article16. Seet CS, Brumbaugh RL, Kee BL. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med. 2004. 199:1689–1700.
Article17. Quong MW, Martensson A, Langerak AW, Rivera RR, Nemazee D, Murre C. Receptor editing and marginal zone B cell development are regulated by the helix-loop-helix protein, E2A. J Exp Med. 2004. 199:1101–1112.
Article18. Ishida D, Kometani K, Yang H, Kakugawa K, Masuda K, Iwai K, Suzuki M, Itohara S, Nakahata T, Hiai H, Kawamoto H, Hattori M, Minato N. Myeloproliferative stem cell disorders by deregulated Rap1 activation in SPA-1-deficient mice. Cancer Cell. 2003. 4:55–65.
Article19. Kurachi H, Wada Y, Tsukamoto N, Maeda M, Kubota H, Hattori M, Iwai K, Minato N. Human SPA-1 gene product selectively expressed in lymphoid tissues is a specific GTPase-activating protein for Rap1 and Rap2. Segregate expression profiles from a rap1GAP gene product. J Biol Chem. 1997. 272:28081–28088.
Article20. Wang SF, Aoki M, Nakashima Y, Shinozuka Y, Tanaka H, Taniwaki M, Hattori M, Minato N. Development of Notch-dependent T-cell leukemia by deregulated Rap1 signaling. Blood. 2008. 111:2878–2886.
Article21. Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006. 6:347–359.
Article22. Weng AP, Ferrando AA, Lee W, Morris JP 4th, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004. 306:269–271.
Article23. Dumortier A, Jeannet R, Kirstetter P, Kleinmann E, Sellars M, dos Santos NR, Thibault C, Barths J, Ghysdael J, Punt JA, Kastner P, Chan S. Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol. 2006. 26:209–220.
Article24. Lin YW, Nichols RA, Letterio JJ, Aplan PD. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood. 2006. 107:2540–2543.
Article25. O'Neil J, Calvo J, McKenna K, Krishanamoorthy V, Aster JC, Bassing CH, Alt FW, Kelliher M, Look AT. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006. 107:781–785.26. O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, Draetta G, Sears R, Clurman BE, Look AT. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007. 204:1813–1824.27. Ishida D, Su L, Tamura A, Katayama Y, Wang S-F, Taniwaki M, Hamazaki Y, Hattori M, Minato N. Rap1 signal controls B cell receptor repertoire and generation of self-reactive B1a cells. Immunity. 2006. 24:417–427.
Article28. Minato N, Hattori M. Spa-1 (Sipa-1) and Rap signaling in leukemia and cancer metastasis. Cancer Sci. 2009. 100:17–23.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Relationship between Recurrent Abdominal Pain and Depressive Trends in School-Aged Children
- Modulation of cartilage differentiation by melanoma inhibiting activity/cartilage-derived retinoic acid-sensitive protein (MIA/CD-RAP)
- Relation between Recurrent Abdominal Pain and Helicobacter pylori Infection and the Role of CagA and VacA in Pediatric Helicobacter pylori Infection
- T-large granular lymphocytic leukemia
- Transmembrane Adaptor Proteins Positively Regulating the Activation of Lymphocytes