J Bacteriol Virol.
2011 Mar;41(1):37-45. 10.4167/jbv.2011.41.1.37.
Genospecies Classification of Acinetobacter calcoaceticus-Acinetobacter baumannii Complex by Restriction Fragment Length Polymorphism
- Affiliations
-
- 1Department of Clinical Laboratory Science, Catholic University of Pusan, Busan, Korea. kschang@cup.ac.kr
- KMID: 1449861
- DOI: http://doi.org/10.4167/jbv.2011.41.1.37
Abstract
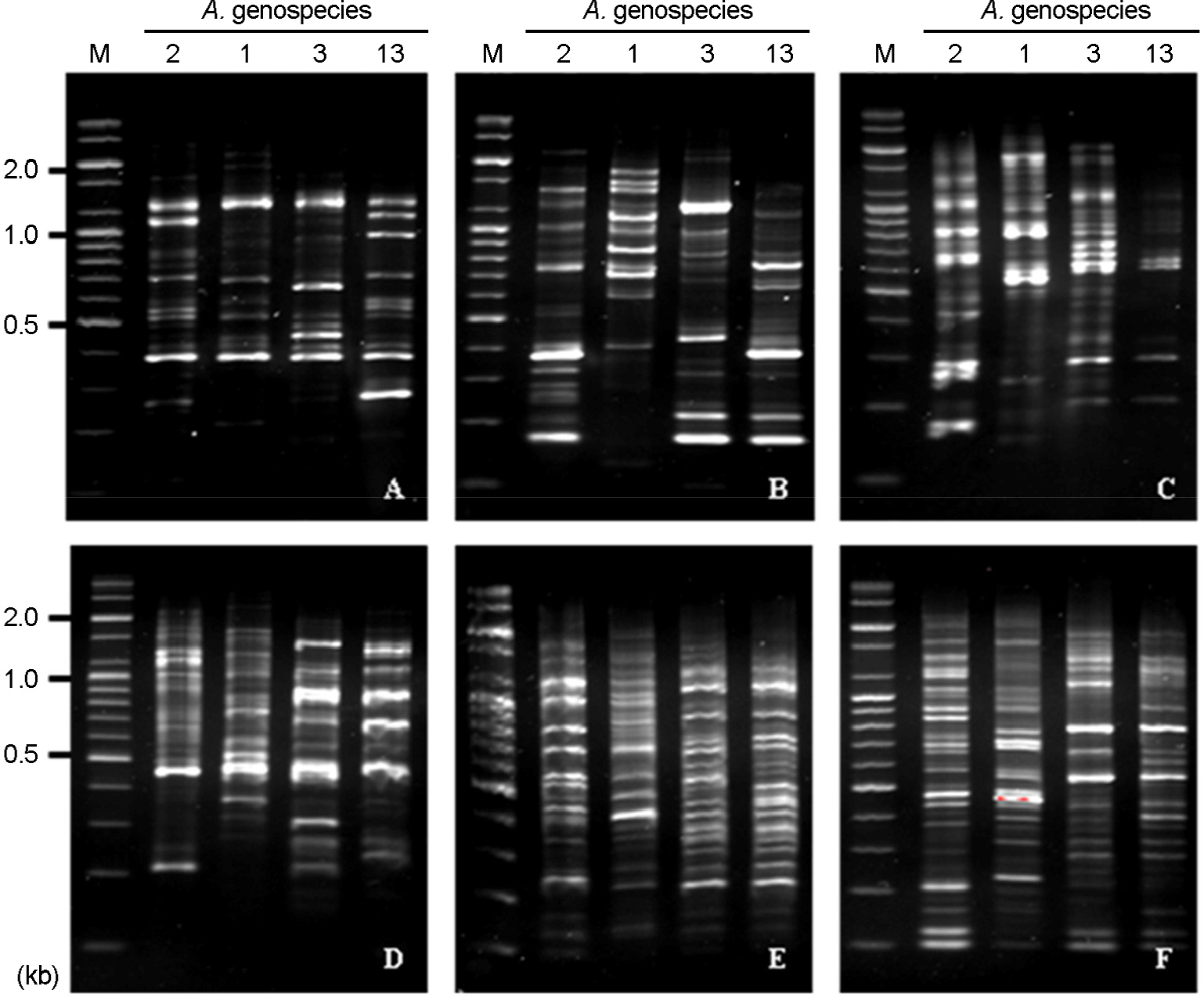
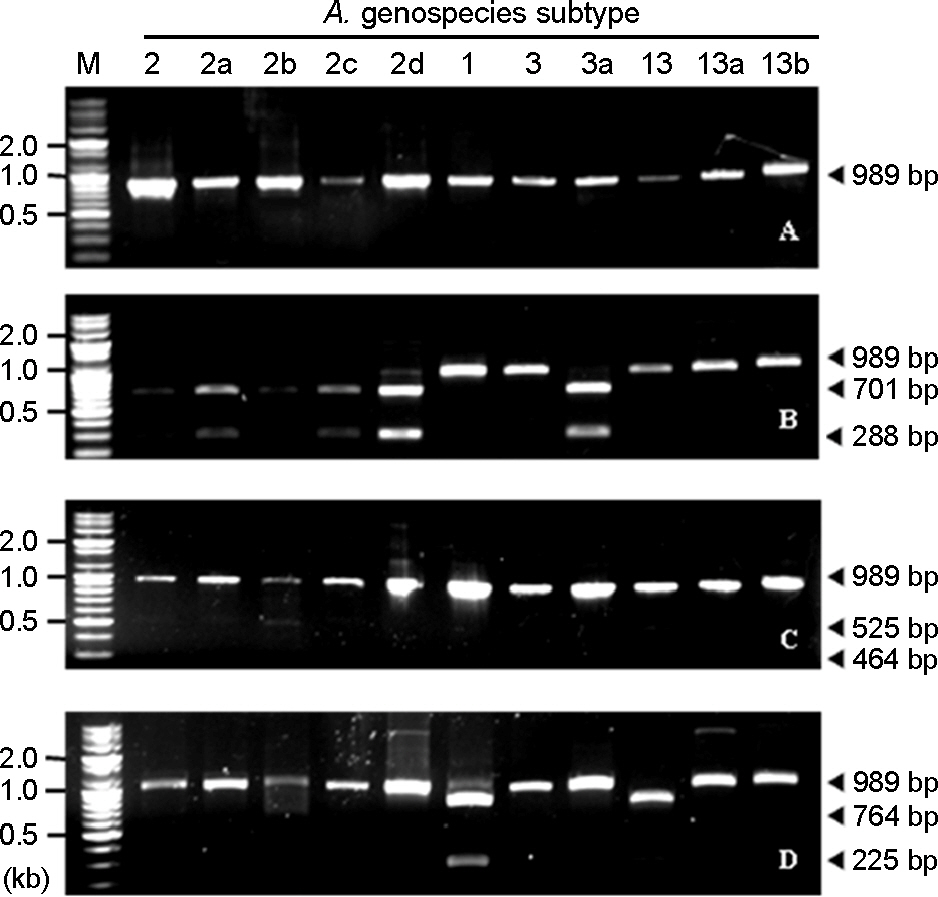
- Acinetobacter calcoaceticus-Acinetobacter baumannii (A. calcoaceticus-A. baumannii) complex, which includes A. calcoaceticus (genospecies 1), A. baumannii (genospecies 2), Acinetobacter genospecies 3 and 13, has been identified as A. baumannii by automated bacteria identification system. The purpose of this study is to develop rapid genospecies classification of A. calcoaceticus-A. baumannii complex by molecular techniques. Random amplified polymorphic DNA (RAPD) and restriction fragment length polymorphism (RFLP) were determined for 4 reference strains and 80 isolates of A. calcoaceticus-A. baumannii complex from clinical sources. Four and eleven RAPD patterns were observed among the reference strains and the isolates, respectively. RAPD might be useful for genomic typing but not for genospecies classification of Acinetobacter spp. RFLP of 16S-23S rRNA intergenic spacer gene with three selected restriction enzymes (ApaLI, SwaI, and SalI) showed only four RFLP patterns in the reference and the isolates. Of 80 isolates, 10 of A. calcoaceticus (12.5%), 50 of A. baumannii (62.5%), 11 of A. genospecies 3 (13.75%), and 9 of A. genospecies 13 (11.25%) were classified by RFLP. This result suggests that RFLP of 16S-23S rRNA intergenic spacer gene of A. calcoaceticus-A. baumannii complex might be useful for genospecies classification.
Keyword
Figure
Reference
-
1). Soddell JA., Beacham AM., Seviour RJ. Phenotypic identification of non-clinical isolates of Acinetobacter species. J Appl Bacteriol. 1993. 74:210–4.2). Dolzani L., Tonin E., Lagatolla C., Prandin L., Monti-Bragadin C. Identification of Acinetobacter isolates in the A. calcoaceticus-A. baumannii complex by restriction analysis of the 16S-23S rRNA intergenic-spacer sequences. J Clin Microbiol. 1995. 33:1108–13.3). Bartual SG., Seifert H., Hippler C., Luzon MA., Wisplinghoff H., Rodríguez-Valera F. Deveplopment of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetoacter baumannii. J Clin Microbiol. 2005. 43:4382–90.4). Vaneechoutte M., Dijkshoorn L., Tjernberg I., Elaichouni A., de Vos P., Claeys G, et al. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J Clin Microbiol. 1995. 33:11–5.5). Ecker JA., Massire C., Hall TA., Ranken R., Pennella TT., Agasino Ivy C, et al. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J Clin Microbiol. 2006. 44:2921–32.6). Bergogne-Bérézin E., Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiolgical, clinical, and epidemiological features. Clin Microbiol Rev. 1996. 9:148–65.7). Koeleman JG., Stoof J., Biesmans DJ., Savelkoul PH., Vandenbroucke-Grauls CM. Comparison of amplified rimosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J Clin Microbiol. 1998. 36:2522–9.8). Lee HK., Kim HJ., Kim YC., Cho SR., Lee WG. Clinical and molecular epidemiologic analysis of a nosocomial outbreak of Acinetobacter baumannii in a neonatal intensive care unit. J Korean Pediatr Soc. 2000. 43:43–8.9). Dueñas Díez AI., Bratos Pérez MA., Eiros Bouza JM., Almaraz Gómez A., Gutiórrez Rodríguez P., Miguel Gómez MA, et al. Susceptibility of the Acinetobacter calcoaceticus-A. baumannii complex to imipenem, meropenem, sulbactam and colistin. Int J Antimicrob Agents. 2004. 23:487–93.10). Dijkshoorn L., Aucken H., Gerner-smidt P., Janssen P., Kaufmann ME., Garaizar J, et al. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotype and phenotypic methods. J Clin Microbiol. 1996. 34:1519–25.11). Chang HC., Wei YF., Dijkshoorn L., Vaneechoutte M., Tang CT., Chang TC. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J Clin Microbiol. 2005. 43:1632–9.12). Bernards AT., Dijkshoorn L., van der Toorn J., Bochner BR., van Boven CP. Phenotypic characterisation of Acinetobacter strains of 13 DNA-DNA hybridization groups by means of the Biolog system. J Med Microbiol. 1995. 42:113–9.13). Lee SH., Lee MK., Park AJ., Choi ES. Identification of Acinetobacter genospecies by analysis of rRNA spacer regions. Korean J Clin Pathol. 2000. 20:48–55.14). Graser Y., Klare I., Halle E., Gantenberg R., Buchholz P., Jacobi HD, et al. Epidemiological study of an Acinetobacter baumannii outbreak by using polymerase chain reaction fingerprinting. J Clin Microbiol. 1993. 31:2417–20.15). Shin MG., Kim SH., Lee JC., Cho D., Kee SJ., Shin JH, et al. A comparison of ID32 GN system with amplified ribosomal DNA restriction analysis for identification of Acinetobacter baumannii. Korean J Lab Med. 2004. 24:107–12.16). Tankovic J., Legrand P., De Gatines G., Chemineau V., Brun-Buisson C., Duval J. Characterization of a hospital outbreak of imipenem-resistant Acinetobacter baumannii by phenotypic and genotypic typing methods. J Clin Microbiol. 1994. 32:2677–81.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Identification of Acinetobacter calcoaceticus - Acinetobacter baumannii Complex by Ribotyping
- Genomic species identification of Acinetobacter calcoaceticus - Acinetobacter baumannii complex strains by amplified ribosomal DNA restriction analysis (ARDRA)
- Identification of Acinetobacter Genospecies by Analysis of rRNA Spacer Regions
- A Comparison of ID 32 GN System with Amplified Ribosomal DNA Restriction Analysis for Identification of Acinetobacter baumannii
- A case of community-acquired acinetobacter calcoaceticus pneumonia