Korean J Lab Med.
2011 Apr;31(2):107-114. 10.3343/kjlm.2011.31.2.107.
Multilocus Sequence Typing for Candida albicans Isolates from Candidemic Patients: Comparison with Southern Blot Hybridization and Pulsed-field Gel Electrophoresis Analysis
- Affiliations
-
- 1Department of Laboratory Medicine, Chonnam National University Medical School, Gwangju, Korea. shinjh@chonnam.ac.kr
- KMID: 1094350
- DOI: http://doi.org/10.3343/kjlm.2011.31.2.107
Abstract
- BACKGROUND
We evaluated the efficacy of multilocus sequence typing (MLST) for assessing the genetic relationship among Candida albicans isolates from patients with candidemia in a hospital setting.
METHODS
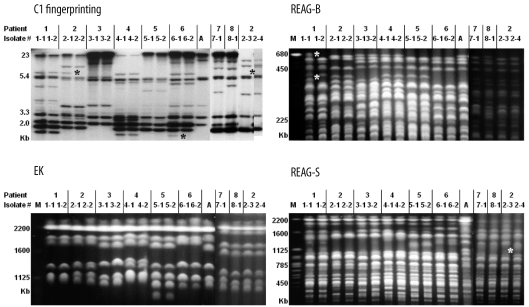
A total of 45 C. albicans isolates from 21 patients with candidemia were analyzed. The MLST results were compared with results obtained by Southern blot hybridization (C1 fingerprinting) and pulsed-field gel electrophoresis (PFGE). PFGE analysis included karyotyping and restriction endonuclease analysis of genomic DNAs using BssHII (REAG-B) and SfiI (REAG-S).
RESULTS
The 45 isolates yielded 20 unique diploid sequence types (DSTs) by MLST, as well as 12 karyotypes, 15 REAG-B patterns, 13 REAG-S patterns, and 14 C1 fingerprinting types. Microevolution among intra-individual isolates was detected in 6, 5, 3, 5, and 7 sets of isolates by MLST (1 or 2 allelic differences), REAG-B, REAG-S, C1 fingerprinting, and a combination of all methods, respectively. Among 20 DSTs, 17 were unique, and 3 were found in more than 1 patient. The results of 2 DSTs obtained from 9 patient isolates were in agreement with REAG and C1 fingerprinting patterns. However, the remaining DST, which was shared by 2 patient isolates, showed 2 different PFGE and C1 fingerprinting patterns. In addition, 3 sets of isolates from different patients, which differed in only 1 or 2 alleles by MLST, also exhibited different PFGE or C1 fingerprinting patterns.
CONCLUSIONS
MLST is highly discriminating among C. albicans isolates, but it may have some limitations in typing isolates from different patients, which may necessitate additional analysis using other techniques.
Keyword
MeSH Terms
Figure
Reference
-
1. Marco F, Lockhart SR, Pfaller MA, Pujol C, Rangel-Frausto MS, Wiblin T, et al. Elucidating the origins of nosocomial infections with Candida albicans by DNA fingerprinting with the complex probe Ca3. J Clin Microbiol. 1999; 37:2817–2828. PMID: 10449459.2. Asmundsdóttir LR, Erlendsdóttir H, Haraldsson G, Guo H, Xu J, Gottfredsson M. Molecular epidemiology of candidemia: evidence of clusters of smoldering nosocomial infections. Clin Infect Dis. 2008; 47:e17–e24. PMID: 18549311.
Article3. Shin JH, Park MR, Song JW, Shin DH, Jung SI, Cho D, et al. Microevolution of Candida albicans strains during catheter-related candidemia. J Clin Microbiol. 2004; 42:4025–4031. PMID: 15364985.4. Lockhart SR, Fritch JJ, Meier AS, Schröppel K, Srikantha T, Galask R, et al. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J Clin Microbiol. 1995; 33:1501–1509. PMID: 7650175.5. Pfaller MA, Lockhart SR, Pujol C, Swails-Wenger JA, Messer SA, Edmond MB, et al. Hospital specificity, region specificity, and fluconazole resistance of Candida albicans bloodstream isolates. J Clin Microbiol. 1998; 36:1518–1529. PMID: 9620370.6. Bougnoux ME, Tavanti A, Bouchier C, Gow NA, Magnier A, Davidson AD, et al. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J Clin Microbiol. 2003; 41:5265–5266. PMID: 14605179.7. Bougnoux ME, Aanensen DM, Morand S, Théraud M, Spratt BG, d'Enfert C. Multilocus sequence typing of Candida albicans: strategies, data exchange and applications. Infect Genet Evol. 2004; 4:243–252. PMID: 15450203.8. Chen KW, Chen YC, Lo HJ, Odds FC, Wang TH, Lin CY, et al. Multilocus sequence typing for analyses of clonality of Candida albicans strains in Taiwan. J Clin Microbiol. 2006; 44:2172–2178. PMID: 16757617.9. Scherer S, Stevens DA. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987; 25:675–679. PMID: 3033016.10. Chowdhary A, Lee-Yang W, Lasker BA, Brandt ME, Warnock DW, Arthington-Skaggs BA. Comparison of multilocus sequence typing and Ca3 fingerprinting for molecular subtyping epidemiologically-related clinical isolates of Candida albicans. Med Mycol. 2006; 44:405–417. PMID: 16882607.11. Odds FC, Bougnoux ME, Shaw DJ, Bain JM, Davidson AD, Diogo D, et al. Molecular phylogenetics of Candida albicans. Eukaryot Cell. 2007; 6:1041–1052. PMID: 17416899.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular Typing in Public Health Laboratories: From an Academic Indulgence to an Infection Control Imperative
- DNA Fingerprinting of Candida albicans Strains Isolated from Candidemic Patients by Polymerase Chain Reaction and Southern Hybridization Methods
- Emergence of optrA-Mediated Linezolid-Nonsusceptible Enterococcus faecalis in a Tertiary Care Hospital
- Molecular Typing of
Mycobacterium intracellulare Using Pulsed-Field Gel Electrophoresis, Variable-Number Tandem-Repeat Analysis, Mycobacteria Interspersed Repetitive-Unit-Variable-Number Tandem Repeat Typing, and Multilocus Sequence Typing: Molecular Characterization and Comparison of Each Typing Methods - Molecular Typing of Candida parapsilosis Isolates from Patients and Healthcare Workers by Pulsed-Field Gel Electrophoresis