Ann Lab Med.
2020 Jul;40(4):321-325. 10.3343/alm.2020.40.4.321.
Emergence of optrA-Mediated Linezolid-Nonsusceptible Enterococcus faecalis in a Tertiary Care Hospital
- Affiliations
-
- 1Department of Laboratory Medicine, University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea. mnkim@amc.seoul.kr
- 2Department of Laboratory Medicine, Inje University Ilsan Paik Hospital, Goyang, Korea.
- KMID: 2470329
- DOI: http://doi.org/10.3343/alm.2020.40.4.321
Abstract
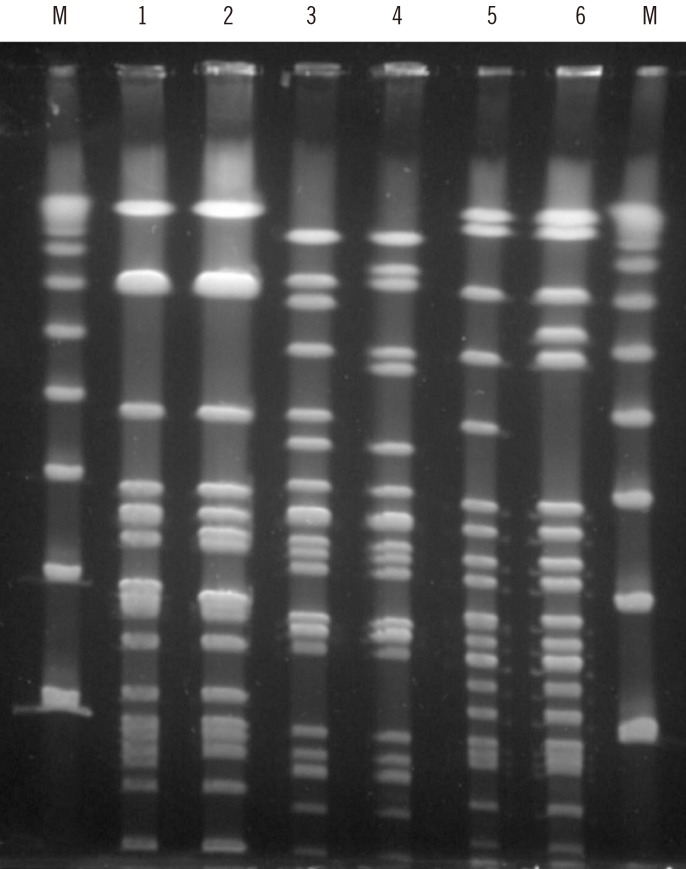
- This study investigated resistance mechanisms and epidemiology of emerging linezolid-nonsusceptible Enterococcus faecalis (LNSEF) in a tertiary care hospital. LNSEF isolated from clinical samples were collected from November 2017 to June 2019. The isolates were investigated for linezolid resistance and the associated molecular mechanisms, including mutations of 23S rRNA domain V and acquisition of the cfr or optrA resistance gene. We used pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing for the molecular typing of the isolates. Among 4,318 E. faecalis isolates, 10 (0.23%) were linezolid-nonsusceptible. All LNSEF isolates were optrA-positive and cfr-negative. Of these isolates, five were sequence type (ST) 476, two ST585, one ST16, one ST16-like, and one ST480. Six LNSEF isolates obtained in the first year clustered to three types in the PFGE analysis: two ST476 isolates of type A, two ST585 isolates of type B, and two ST16 or ST16-like isolates of type C. Seven cases were of community-onset and three were hospital acquired, but total of eight were healthcare-associated including five community-onset. None of the patients had a history of linezolid treatment, and in one patient, we detected linezolid-susceptible E. faecalis one month before LNSEF detection. In conclusion, heterogenous clones of optrA-positive LNSEF emerged in the hospital mainly via community-onset.
Keyword
MeSH Terms
Figure
Reference
-
1. Mendes RE, Deshpande LM, Jones RN. Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist Updat. 2014; 17:1–12. PMID: 24880801.2. Bae HG, Sung H, Kim MN, Lee EJ, Koo Lee S. First report of a linezolid- and vancomycin-resistant Enterococcus faecium strain in Korea. Scand J Infect Dis. 2006; 38:383–386. PMID: 16709544.3. Lee SM, Huh HJ, Song DJ, Shim HJ, Park KS, Kang CI, et al. Resistance mechanisms of linezolid-nonsusceptible enterococci in Korea: low rate of 23S rRNA mutations in Enterococcus faecium. J Med Microbiol. 2017; 66:1730–1735. PMID: 29111969.4. Cui L, Wang Y, Lv Y, Wang S, Song Y, Li Y, et al. Nationwide surveillance of novel oxazolidinone resistance gene optrA in Enterococcus isolates in China from 2004 to 2014. Antimicrob Agents Chemother. 2016; 60:7490–7493. PMID: 27645239.5. Deshpande LM, Castanheira M, Flamm RK, Mendes RE. Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: results from the SENTRY Antimicrobial Surveillance Program. J Antimicrob Chemother. 2018; 73:2314–2322. PMID: 29878213.6. Hua R, Xia Y, Wu W, Yang M, Yan J. Molecular epidemiology and mechanisms of 43 low-level linezolid-resistant Enterococcus faecalis strains in Chongqing, China. Ann Lab Med. 2019; 39:36–42. PMID: 30215228.7. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995; 33:2233–2239. PMID: 7494007.8. Pfaller MA, Mendes RE, Streit JM, Hogan PA, Flamm RK. Five-year summary of in vitro activity and resistance mechanisms of linezolid against clinically important Gram-positive cocci in the United States from the LEADER surveillance program (2011 to 2015). Antimicrob Agents Chemother. 2017; 61:e00609–e00617. PMID: 28483950.9. Jung J, Park K, Shin SH, Lee JY, Kim MN, Kim SH. The pitfall of cohort isolation in an outbreak of linezolid-resistant, vancomycin-resistant enterococci. Clin Microbiol Infect. 2019; 25:1568–1569. PMID: 31449869.10. Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z, et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother. 2015; 70:2182–2190. PMID: 25977397.11. Zhang Y, Dong G, Li J, Chen L, Liu H, Bi W, et al. A high incidence and coexistence of multiresistance genes cfr and optrA among linezolid-resistant enterococci isolated from a teaching hospital in Wenzhou, China. Eur J Clin Microbiol Infect Dis. 2018; 37:1441–1448. PMID: 29909468.12. Zhou W, Gao S, Xu H, Zhang Z, Chen F, Shen H, et al. Distribution of the optrA gene in Enterococcus isolates at a tertiary care hospital in China. J Glob Antimicrob Resist. 2019; 17:180–186. PMID: 30641287.13. Bender JK, Fleige C, Lange D, Klare I, Werner G. Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int J Antimicrob Agents. 2018; 52:819–827. PMID: 30236952.14. Argudín MA, Youzaga S, Dodémont M, Heinrichs A, Roisin S, Deplano A, et al. Detection of optrA-positive enterococci clinical isolates in Belgium. Eur J Clin Microbiol Infect Dis. 2019; 38:985–987. PMID: 30771123.15. Sassi M, Guérin F, Zouari A, Beyrouthy R, Auzou M, Fines-Guyon M, et al. Emergence of optrA-mediated linezolid resistance in enterococci from France, 2006-16. J Antimicrob Chemother. 2019; 74:1469–1472. PMID: 30897199.16. Cai J, Schwarz S, Chi D, Wang Z, Zhang R, Wang Y. Faecal carriage of optrA-positive enterococci in asymptomatic healthy humans in Hangzhou, China. Clin Microbiol Infect. 2019; 25:630.e1–630.e6. PMID: 30076974.17. Na SH, Moon DC, Choi MJ, Oh SJ, Jung DY, Kang HY, et al. Detection of oxazolidinone and phenicol resistant enterococcal isolates from duck feces and carcasses. Int J Food Microbiol. 2019; 293:53–59. PMID: 30640000.18. Tamang MD, Moon DC, Kim SR, Kang HY, Lee K, Nam HM, et al. Detection of novel oxazolidinone and phenicol resistance gene optrA in enterococcal isolates from food animals and animal carcasses. Vet Microbiol. 2017; 201:252–256. PMID: 28284617.19. Huang J, Chen L, Wu Z, Wang L. Retrospective analysis of genome sequences revealed the wide dissemination of optrA in Gram-positive bacteria. J Antimicrob Chemother. 2017; 72:614–616. PMID: 27999025.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular Epidemiology and Mechanisms of 43 Low-Level Linezolid-Resistant Enterococcus faecalis Strains in Chongqing, China
- Susceptibility of Glycopeptide-Resistant Enterococci to Linezolid, Quinupristin/dalfopristin, Tigecycline and Daptomycin in a Tertiary Greek Hospital
- A Case of Bilateral Endogenous Enterococcus Faecalis Endophthalmitis in Liver Abscess
- Linezolid-Resistant Coagulase-Negative Staphylococci in a Tertiary Hospital: Molecular Epidemiology, Clinical Characteristics, and Outcomes
- High Incidence of Virulence Factors Among Clinical Enterococcus faecalis Isolates in Southwestern Iran