J Vet Sci.
2006 Sep;7(3):277-280. 10.4142/jvs.2006.7.3.277.
The 23S rRNA gene PCR-RFLP used for characterization of porcine intestinal spirochete isolates
- Affiliations
-
- 1School of Veterinary and Biomedical Sciences, Murdoch University, Murdoch, WA 6150, Australia.
- 2College of Veterinary Medicine, Chonnam National University, Gwangju 500-757, Korea. jaeil@chonnam.ac.kr
- KMID: 1089913
- DOI: http://doi.org/10.4142/jvs.2006.7.3.277
Abstract
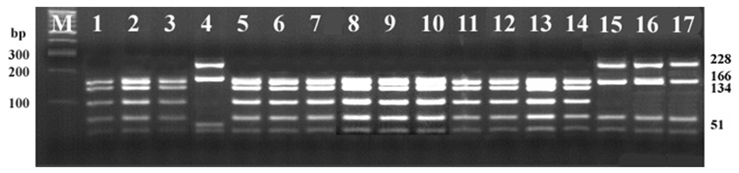
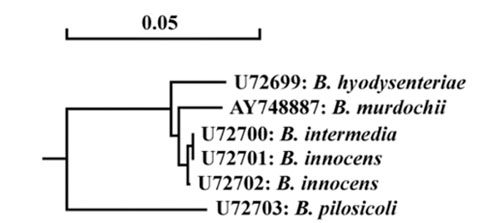
- Using three reference strains of Brachyspira hyodysenteriae (B204, B234, B169), one B. pilosicoli (P43/6/78), one B. murdochii (56-150), one B. intermedia (PWS/A), one B. innocens (B256) and ten Korean isolates, PCR-RFLP analysis of DNA encoding 23S rRNA was performed to establish a rapid and accurate method for characterizing porcine intestinal spirochetes. Consequently, B. hyodysenteriae and B. pilosicoli revealed different restriction patterns; however, the other three species shared the same pattern. These findings are not consistent with a prior report. Differences in 23S rRNA gene sequences, between two B. murdochii strains, 56-150 and 155-20, were observed. These results indicate that 23S rRNA PCR-RFLP could be used as an identification method for pathogenic Brachyspira spp. (B. hyodysenteriae and B. pilosicoli) as well as an epidemiological tool for characterizing spirochetes isolated from swine.
Keyword
MeSH Terms
-
Animals
DNA, Bacterial/genetics
Dysentery, Bacillary/diagnosis/microbiology/*veterinary
Korea
Phylogeny
Polymerase Chain Reaction/methods/*veterinary
Polymorphism, Restriction Fragment Length
RNA, Ribosomal, 23S/chemistry/*genetics
Spirochaetales/*genetics/*isolation&purification
Spirochaetales Infections/diagnosis/microbiology/*veterinary
Swine
Swine Diseases/diagnosis/*microbiology
Figure
Reference
-
1. Atyeo RF, Oxberry SL, Combs BG, Hampson DJ. Development and evaluation of polymerase chain reaction tests as an aid to diagnosis of swine dysentery and intestinal spirochaetosis. Lett Appl Microbiol. 1998. 26:126–130.
Article2. Atyeo RF, Stanton TB, Jensen NS, Suriyaarachichi DS, Hampson DJ. Differentiation of Serpulina species by NADH oxidase gene (nox) sequence comparisons and nox-based polymerase chain reaction tests. Vet Microbiol. 1999. 67:47–60.
Article3. Barcellos DESN, de Uzeda M, Ikuta N, Lunge VR, Fonseca AS, Kader II, Duhamel GE. Identification of porcine intestinal spirochetes by PCR-restriction fragment length polymorphism analysis of ribosomal DNA encoding 23S rRNA. Vet Microbiol. 2000. 75:189–198.
Article4. De Smet KA, Worth DE, Barrett SP. Variation amongst human isolates of Brachyspira (Serpulina) pilosicoli based on biochemial characterization and 16S rRNA gene sequencing. Int J Syst Bacteriol. 1998. 48:1257–1263.
Article5. Dugourd D, Jacques M, Bigras-Poulin M, Harel J. Characterization of Serpulina hyodysenteriae isolates of serotypes 8 and 9 by random amplification of polymorphic DNA analysis. Vet Microbiol. 1996. 48:305–314.
Article6. Duhamel GE, Muniappa N, Mathiesen MR, Johnson JL, Toth J, Elder RO, Doster AR. Certain weakly beta-haemolytic intestinal spirochetes are phenotypically and genotypically related to spirochetes associated with human and porcine intestinal spirochaetosis. J Clin Microbiol. 1995. 33:2212–2215.
Article7. Elder RO, Duhamel GE, Mathiesen MR, Erickson ED, Gebhart CJ, Oberst RD. Multiplex polymerase chain reaction for simultaneous detection of Lawsonia intracellularis, Serpulina hyodysenteriae, and salmonellae in porcine intestinal specimens. J Vet Diagn Invest. 1997. 9:281–286.
Article8. Fisher LN, Mathiesen MR, Duhamel GE. Restriction fragment length polymorphism of the periplasmic flagellar flaA1 gene of Serpulina species. Clin Diagn Lab Immunol. 1997. 4:681–686.
Article9. Hampson DJ. Slide-agglutination for rapid serological typing of Treponema hyodysenteriae. Epidemiol Infect. 1991. 106:541–547.
Article10. Kim TJ, Jung SC, Lee JI. Characterization of Brachyspira hyodysenteriae isolates from Korea. J Vet Sci. 2005. 6:335–339.
Article11. Koopman MB, Kasbohrer A, Beckmann G, van der Zeijst BA, Kusters JG. Genetic similarity of intestinal spirochetes from humans and various animal species. J Clin Microbiol. 1993. 31:711–716.
Article12. Lee JI, Hampson DJ. Genetic characterisation of intestinal spirochetes and their association with disease. J Med Microbiol. 1994. 40:365–371.
Article13. Lee JI, Hampson DJ, Lymbery AJ, Harders SJ. The porcine intestinal spirochetes: identification of new genetic groups. Vet Microbiol. 1993. 34:273–285.
Article14. Leser TD, Moller K, Jensen TK, Jorsal SE. Specific detection of Serpulina hyodysenteriae and potentially pathogenic weakly beta-haemolytic porcine intestinal spirochetes by polymerase chain reaction targeting 23S rDNA. Mol Cell Probes. 1997. 11:363–372.
Article15. Muniappa N, Mathiesen MR, Duhamel GE. Laboratory identification and enteropathogenicity testing of Serpulina pilosicoli associated with porcine colonic spirochetosis. J Vet Diagn Invest. 1997. 9:165–171.
Article16. Rohde J, Rothkamp A, Gerlach GF. Differentiation of porcine Brachyspira species by a novel nox PCR-based restriction fragment length polymorphism analysis. J Clin Microbiol. 2002. 40:2598–2600.
Article17. Stanton TB, Fournie-Amazouz E, Postic D, Trott DJ, Grimont PA, Baranton G, Hampson DJ, Saint Girons I. Recognition of two new species of intestinal spirochetes: Serpulina intermedia sp. nov. and Serpulina murdochii sp. nov. Int J Syst Bacteriol. 1997. 47:1007–1012.
Article18. ter Huurne AA, van Houten M, Koopman MB, van der Zeijst BA, Gaastra W. Characterization of Dutch porcine Serpulina (Treponema) isolates by restriction endonuclease analysis and DNA hybridization. J Gen Microbiol. 1992. 138:1929–1934.19. Townsend KM, Giang VN, Stephens C, Scott PT, Trott DJ. Application of nox-restriction fragment length polymorphism for the differentiation of Brachyspira intestinal spirochetes isolated from pigs and poultry in Australia. J Vet Diagn Invest. 2005. 17:103–109.
Article20. Trott DJ, Stanton TB, Jensen NS, Duhamel GE, Johnson JL, Hampson DJ. Serpulina pilosicoli sp. nov., the agent of porcine intestinal spirochetosis. Int J Syst Bacteriol. 1996. 46:206–215.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genospecies Classification of Acinetobacter calcoaceticus-Acinetobacter baumannii Complex by Restriction Fragment Length Polymorphism
- Classification of Borrelia burgdorferi Sensu Lato Isolated in Korea by Restriction Fragment Length Polymorphism
- Mutations in the 23S rRNA Gene of Helicobacter pylori Associated with Clarithromycin Resistance
- Analysis of Clarithromycin Resistance of Helicobacter pylori Isolated in Korea
- Detection of 23S rRNA Mutation Associated with Clarithromycin Resistance in Children with Helicobacter pylori Infection