J Korean Med Sci.
2024 May;39(17):e157. 10.3346/jkms.2024.39.e157.
Rapid Direct Identification of Microbial Pathogens and Antimicrobial Resistance Genes in Positive Blood Cultures Using a Fully Automated Multiplex PCR Assay
- Affiliations
-
- 1Department of Laboratory Medicine, Korea University College of Medicine, Seoul, Korea
- KMID: 2555498
- DOI: http://doi.org/10.3346/jkms.2024.39.e157
Abstract
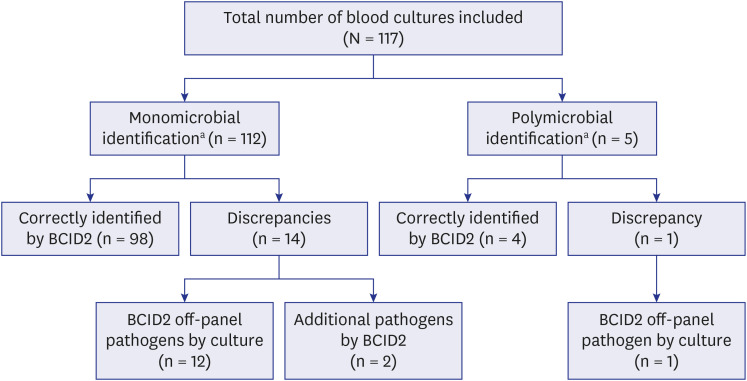
- This study assessed the performance of the BioFire Blood Culture Identification 2 (BCID2) panel in identifying microorganisms and antimicrobial resistance (AMR) profiles in positive blood cultures (BCs) and its influence on turnaround time (TAT) compared with conventional culture methods. We obtained 117 positive BCs, of these, 102 (87.2%) were correctly identified using BCID2. The discordance was due to off-panel pathogens detected by culture (n = 13), and additional pathogens identified by BCID2 (n = 2). On-panel pathogen concordance between the conventional culture and BCID2 methods was 98.1% (102/104). The conventional method detected 19 carbapenemase-producing organisms, 14 extendedspectrum beta-lactamase-producing Enterobacterales, 18 methicillin-resistant Staphylococcus spp., and four vancomycin-resistant Enterococcus faecium. BCID2 correctly predicted 53 (96.4%) of 55 phenotypic resistance patterns by detecting AMR genes. The TAT for BCID2 was significantly lower than that for the conventional method. BCID2 rapidly identifies pathogens and AMR genes in positive BCs.
Keyword
Figure
Reference
-
1. McNamara JF, Righi E, Wright H, Hartel GF, Harris PN, Paterson DL. Long-term morbidity and mortality following bloodstream infection: a systematic literature review. J Infect. 2018; 77(1):1–8. PMID: 29746948.2. Son JS, Song JH, Ko KS, Yeom JS, Ki HK, Kim SW, et al. Bloodstream infections and clinical significance of healthcare-associated bacteremia: a multicenter surveillance study in Korean hospitals. J Korean Med Sci. 2010; 25(7):992–998. PMID: 20592888.3. Kim HJ, Oh DK, Lim SY, Cho YJ, Park S, Suh GY, et al. Antibiogram of multidrug-resistant bacteria based on sepsis onset location in Korea: a multicenter cohort study. J Korean Med Sci. 2023; 38(10):e75. PMID: 36918029.4. Goto M, Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect. 2013; 19(6):501–509. PMID: 23473333.5. Prod’hom G, Bizzini A, Durussel C, Bille J, Greub G. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for direct bacterial identification from positive blood culture pellets. J Clin Microbiol. 2010; 48(4):1481–1483. PMID: 20164269.6. Cheong HS, Park KH, Kim HB, Kim SW, Kim B, Moon C, et al. Core elements for implementing antimicrobial stewardship programs in Korean general hospitals. Infect Chemother. 2022; 54(4):637–673. PMID: 36596679.7. Park JM, Kwon M, Hong KH, Lee H, Yong D. European committee on antimicrobial susceptibility testing-recommended rapid antimicrobial susceptibility testing of Escherichia coli, Klebsiella pneumoniae, and Staphylococcus aureus from positive blood culture bottles. Ann Lab Med. 2023; 43(5):443–450. PMID: 37080745.8. Kim KJ, Yun SG, Cho Y, Nam MH, Ko YJ, Lee CK. Evaluation of a sterile, filter-based, in-house method for rapid direct bacterial identification and antimicrobial susceptibility testing using positive blood culture. Eur J Clin Microbiol Infect Dis. 2023; 42(6):691–700. PMID: 37012540.9. Grohs P, Remaud E, Lath C, Vuong K, Parolini ML, Dannaoui E, et al. Comparison of the new VITEK MS PRIME system with the matrix-assisted laser desorption ionization biotyper microflex LT for the identification of microorganisms. Ann Lab Med. 2023; 43(6):574–584. PMID: 37387490.10. Juttukonda LJ, Katz S, Gillon J, Schmitz J, Banerjee R. Impact of a rapid blood culture diagnostic test in a children’s hospital depends on gram-positive versus gram-negative organism and day versus night shift. J Clin Microbiol. 2020; 58(4):e01400-19.11. Banerjee R, Komarow L, Virk A, Rajapakse N, Schuetz AN, Dylla B, et al. Randomized trial evaluating clinical impact of RAPid IDentification and susceptibility testing for gram-negative bacteremia: RAPIDS-GN. Clin Infect Dis. 2021; 73(1):e39–e46. PMID: 32374822.12. Kim D, Lee H, Choi JS, Croney CM, Park KS, Park HJ, et al. The changes in epidemiology of imipenem-resistant Acinetobacter baumannii bacteremia in a pediatric intensive care unit for 17 years. J Korean Med Sci. 2022; 37(24):e196. PMID: 35726147.13. Widyasari K, Lee S, Cho OH, Hong SI, Ryu BH, Kim S. The significance of filmarray blood culture identification panel (FA-BCID) for managing patients with positive blood cultures. Diagnostics (Basel). 2023; 13(21):3335. PMID: 37958231.14. Peker N, Couto N, Sinha B, Rossen JW. Diagnosis of bloodstream infections from positive blood cultures and directly from blood samples: recent developments in molecular approaches. Clin Microbiol Infect. 2018; 24(9):944–955. PMID: 29787889.15. Cortazzo V, D’Inzeo T, Giordano L, Menchinelli G, Liotti FM, Fiori B, et al. Comparing BioFire FilmArray BCID2 and BCID panels for direct detection of bacterial pathogens and antimicrobial resistance genes from positive blood cultures. J Clin Microbiol. 2021; 59(4):e03163-20. PMID: 33472903.16. Jonasson E, Matuschek E, Kahlmeter G. The EUCAST rapid disc diffusion method for antimicrobial susceptibility testing directly from positive blood culture bottles. J Antimicrob Chemother. 2020; 75(4):968–978. PMID: 32016342.17. Tabak YP, Vankeepuram L, Ye G, Jeffers K, Gupta V, Murray PR. Blood culture turnaround time in U.S. acute care hospitals and implications for laboratory process optimization. J Clin Microbiol. 2018; 56(12):e00500-18. PMID: 30135230.18. Banerjee R, Humphries R. Rapid antimicrobial susceptibility testing methods for blood cultures and their clinical impact. Front Med (Lausanne). 2021; 8:635831. PMID: 33777978.19. Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis. 2017; 64(1):15–23. PMID: 27678085.20. Berinson B, Both A, Berneking L, Christner M, Lütgehetmann M, Aepfelbacher M, et al. Usefulness of BioFire FilmArray BCID2 for blood culture processing in clinical practice. J Clin Microbiol. 2021; 59(8):e0054321. PMID: 33980648.21. Holma T, Torvikoski J, Friberg N, Nevalainen A, Tarkka E, Antikainen J, et al. Rapid molecular detection of pathogenic microorganisms and antimicrobial resistance markers in blood cultures: evaluation and utility of the next-generation FilmArray Blood Culture Identification 2 panel. Eur J Clin Microbiol Infect Dis. 2022; 41(3):363–371. PMID: 34350523.22. Graff KE, Palmer C, Anarestani T, Velasquez D, Hamilton S, Pretty K, et al. Clinical impact of the expanded BioFire Blood Culture Identification 2 panel in a U.S. children’s hospital. Microbiol Spectr. 2021; 9(1):e0042921. PMID: 34431685.23. Sparks R, Balgahom R, Janto C, Polkinghorne A, Branley J. Evaluation of the BioFire Blood Culture Identification 2 panel and impact on patient management and antimicrobial stewardship. Pathology. 2021; 53(7):889–895. PMID: 34120744.24. Sze DT, Lau CC, Chan TM, Ma ES, Tang BS. Comparison of novel rapid diagnostic of blood culture identification and antimicrobial susceptibility testing by Accelerate Pheno system and BioFire FilmArray Blood Culture Identification and BioFire FilmArray Blood Culture Identification 2 panels. BMC Microbiol. 2021; 21(1):350. PMID: 34922463.25. Peri AM, Bauer MJ, Bergh H, Butkiewicz D, Paterson DL, Harris PN. Performance of the BioFire Blood Culture Identification 2 panel for the diagnosis of bloodstream infections. Heliyon (Lond). 2022; 8(7):e09983.26. Rhoads DD, Pournaras S, Leber A, Balada-Llasat JM, Harrington A, Sambri V, et al. Multicenter evaluation of the Biofire Blood Culture Identification 2 panel for detection of bacteria, yeasts, and antimicrobial resistance genes in positive blood culture samples. J Clin Microbiol. 2023; 61(6):e0189122. PMID: 37227281.27. Kim J, Lim YM. Prevalence of derepressed ampC mutants and extended-spectrum beta-lactamase producers among clinical isolates of Citrobacter freundii, Enterobacter spp., and Serratia marcescens in Korea: dissemination of CTX-M-3, TEM-52, and SHV-12. J Clin Microbiol. 2005; 43(5):2452–2455. PMID: 15872281.28. Conceição T, Coelho C, de Lencastre H, Aires-de-Sousa M. Frequent occurrence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) strains in two African countries. J Antimicrob Chemother. 2015; 70(12):3200–3204. PMID: 26318189.29. Chen FJ, Huang IW, Wang CH, Chen PC, Wang HY, Lai JF, et al. mecA-positive Staphylococcus aureus with low-level oxacillin MIC in Taiwan. J Clin Microbiol. 2012; 50(5):1679–1683. PMID: 22378906.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Multiplex PCR Assay for Identification of Mycobacterial Species Isolated from Liquid Cultures
- Multiplex PCR analysis of virulence genes and their influence on antibiotic resistance in Enterococcus spp. isolated from broiler chicken
- Evaluation of Seeplex(TM) Pneumobacter Multiplex PCR Kit for the Detection of Respiratory Bacterial Pathogens in Pediatric Patients
- Genomic Characteristics and Identification of Salmonella enterica serovars Typhi and Paratyphi A Using Multiplex PCR
- Evaluation of Vancomycin Resistance 3 Multiplexed PCR Assay for Detection of Vancomycin-Resistant Enterococci from Rectal Swabs