Cancer Res Treat.
2024 Apr;56(2):538-548. 10.4143/crt.2023.1059.
Genomic Characteristics and Its Therapeutic Implications in Breast Cancer Patients with Detectable Molecular Residual Disease
- Affiliations
-
- 1Department of Breast and Thyroid Surgery, Daping Hospital, Army Military Medical University, Chongqing, China
- 2Geneplus-Beijing, Beijing, China
- KMID: 2554343
- DOI: http://doi.org/10.4143/crt.2023.1059
Abstract
- Purpose
Molecular residual disease (MRD) is the main cause of postoperative recurrence of breast cancer. However, the baseline tumor genomic characteristics and therapeutic implications of breast cancer patients with detectable MRD after surgery are still unknown.
Materials and Methods
In this study, we enrolled 80 patients with breast cancer who underwent next-generation sequencing-based genetic testing of 1,021 cancer-related genes performed on baseline tumor and postoperative plasma, among which 18 patients had detectable MRD after surgery.
Results
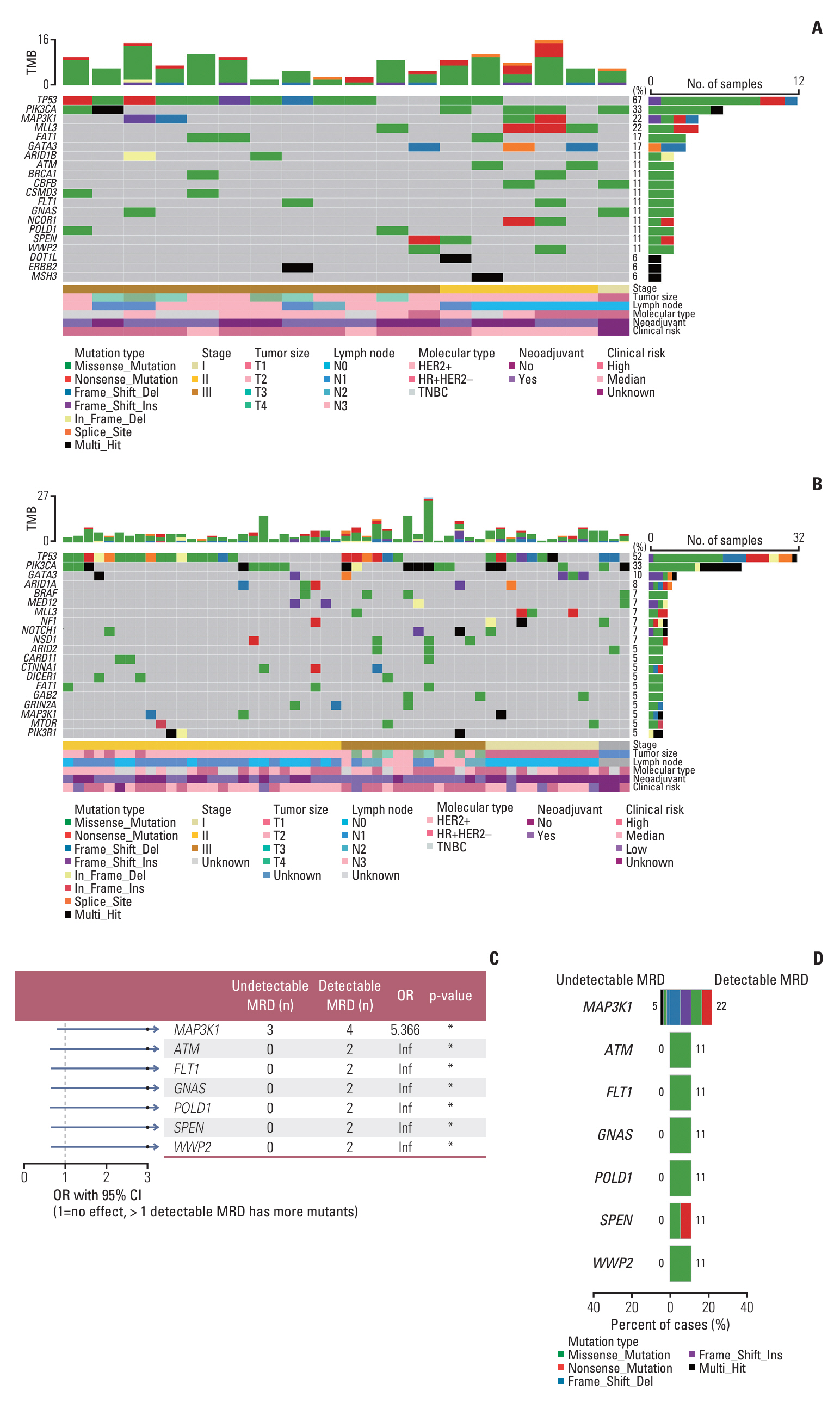
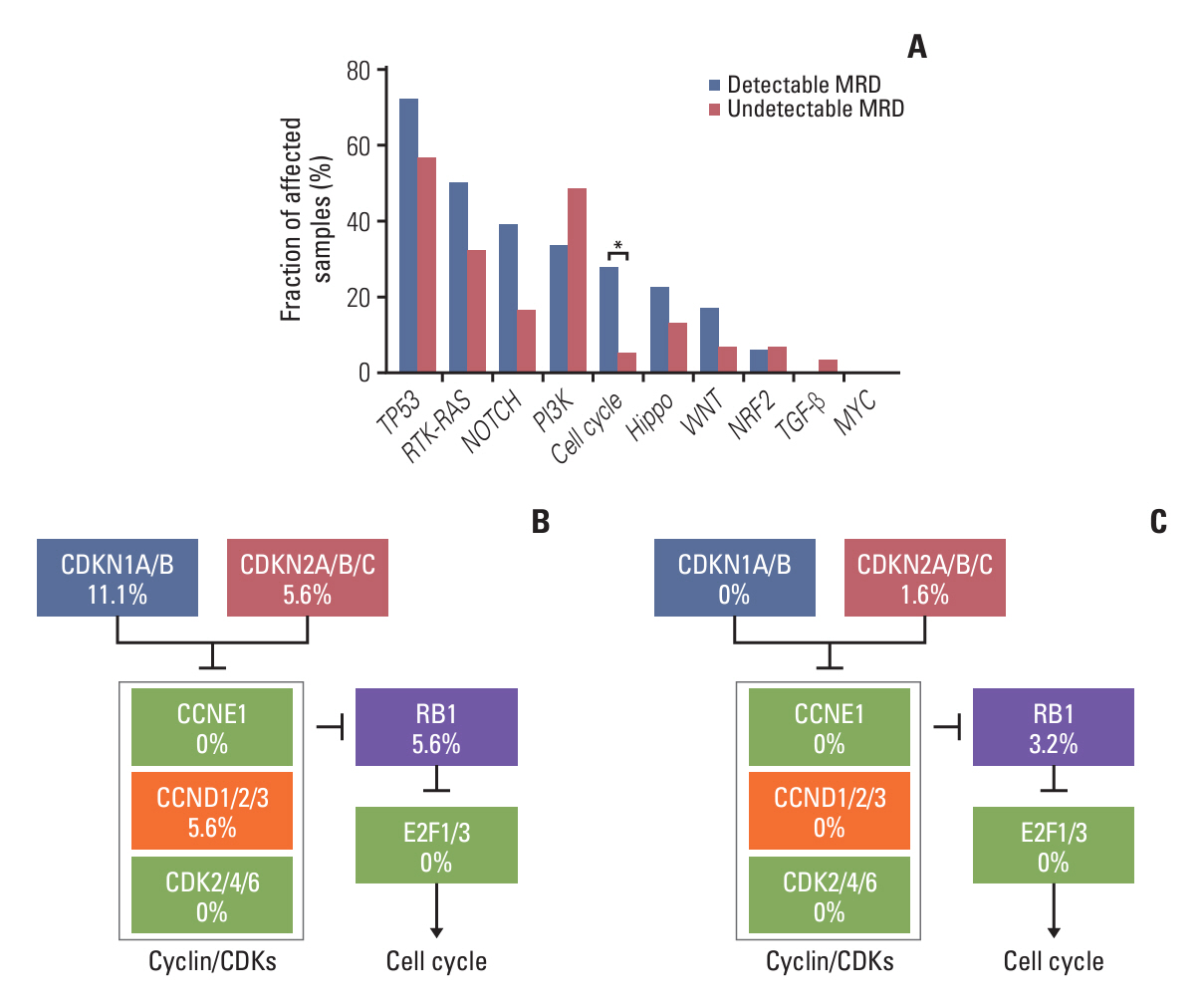
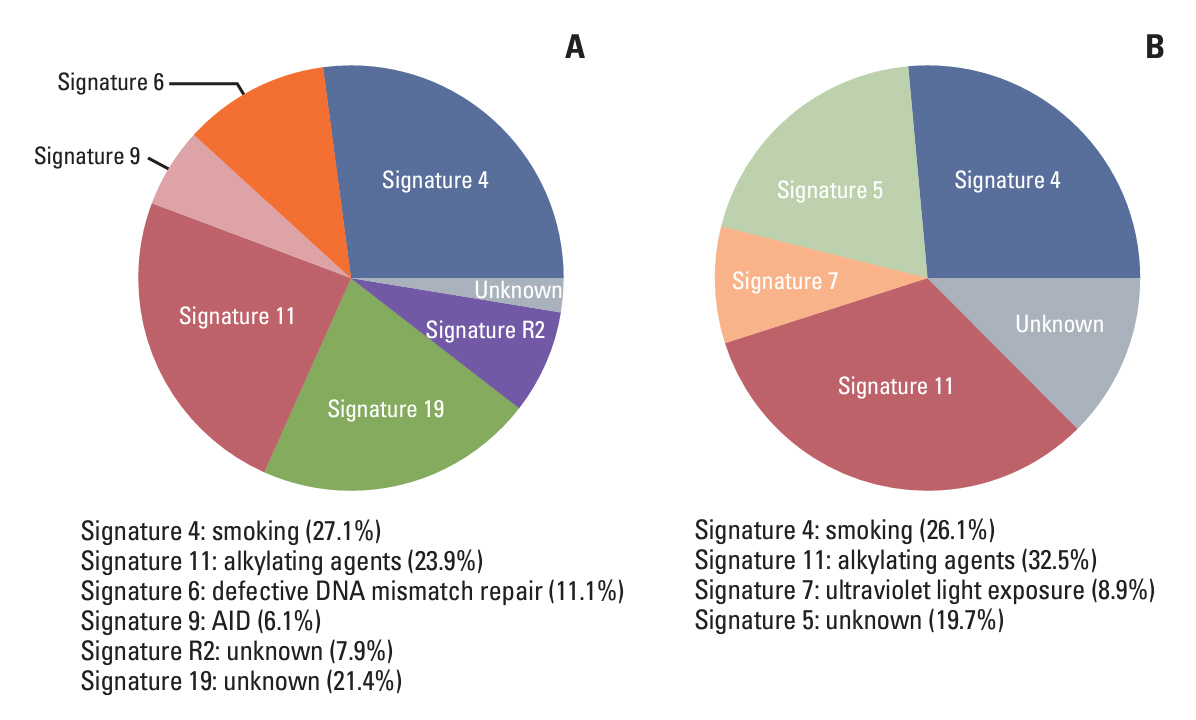
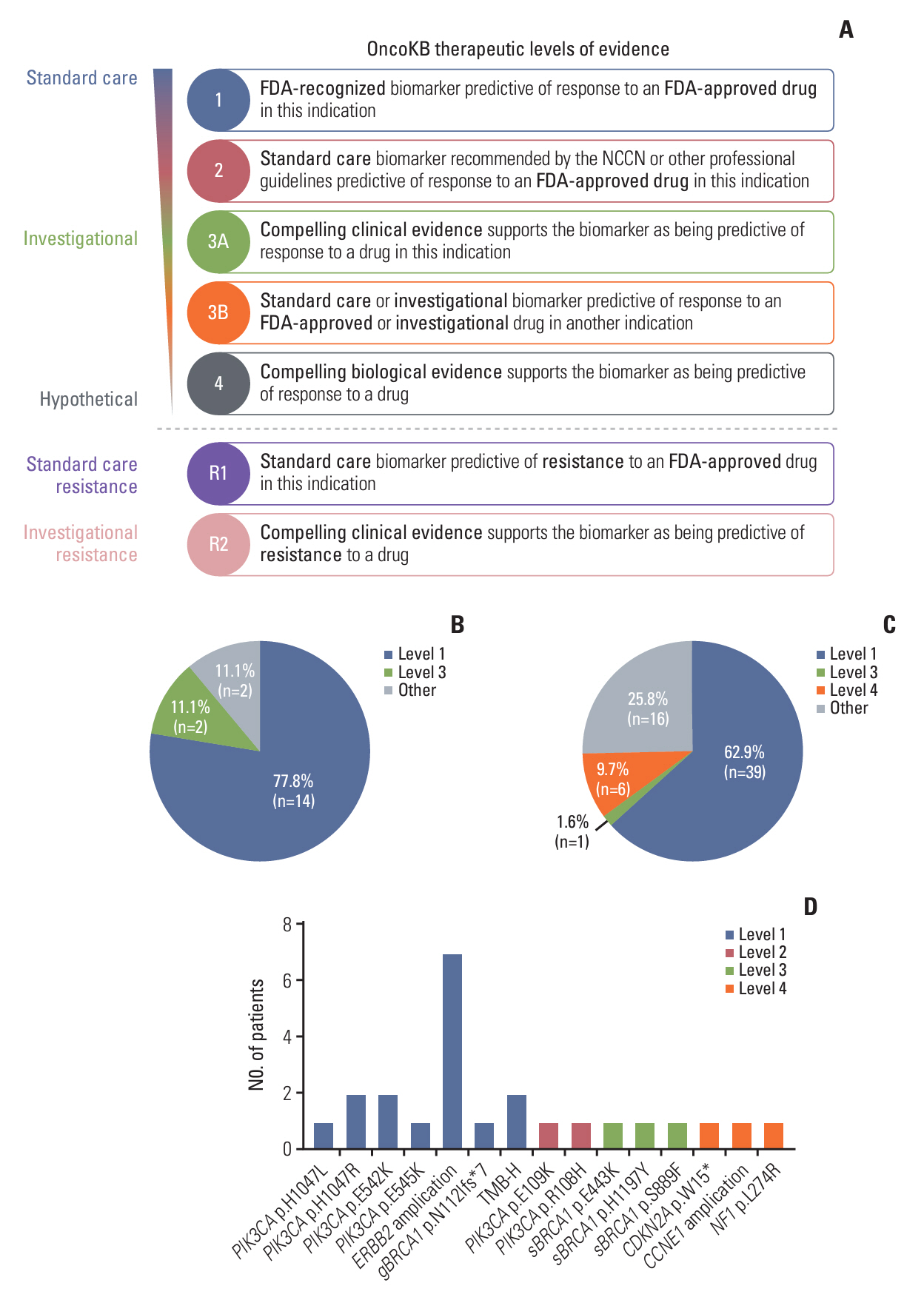
Baseline clinical characteristics found that patients with higher clinical stages were more likely to have detectable MRD. Analysis of single nucleotide variations and small insertions/deletions in baseline tumors showed that somatic mutations in MAP3K1, ATM, FLT1, GNAS, POLD1, SPEN, and WWP2 were significantly enriched in patients with detectable MRD. Oncogenic signaling pathway analysis revealed that alteration of the Cell cycle pathway was more likely to occur in patients with detectable MRD (p=0.012). Mutational signature analysis showed that defective DNA mismatch repair and activation-induced cytidine deaminase (AID) mediated somatic hypermutation (SHM) were associated with detectable MRD. According to the OncoKB database, 77.8% (14/18) of patients with detectable MRD had U.S. Food and Drug Administration–approved mutational biomarkers and targeted therapy.
Conclusion
Our study reports genomic characteristics of breast cancer patients with detectable MRD. The cell cycle pathway, defective DNA mismatch repair, and AID-mediated SHM were found to be the possible causes of detectable MRD. We also found the vast majority of patients with detectable MRD have the opportunity to access targeted therapy.
Keyword
Figure
Reference
-
References
1. Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. Lancet. 2021; 397:1750–69.
Article2. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010; 363:1938–48.
Article3. Coombes RC, Page K, Salari R, Hastings RK, Armstrong A, Ahmed S, et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin Cancer Res. 2019; 25:4255–63.
Article4. Garcia-Murillas I, Chopra N, Comino-Mendez I, Beaney M, Tovey H, Cutts RJ, et al. Assessment of molecular relapse detection in early-stage breast cancer. JAMA Oncol. 2019; 5:1473–8.
Article5. Parsons HA, Rhoades J, Reed SC, Gydush G, Ram P, Exman P, et al. Sensitive detection of minimal residual disease in patients treated for early-stage breast cancer. Clin Cancer Res. 2020; 26:2556–64.
Article6. Lipsyc-Sharf M, de Bruin EC, Santos K, McEwen R, Stetson D, Patel A, et al. Circulating tumor DNA and late recurrence in high-risk hormone receptor-positive, human epidermal growth factor receptor 2-negative breast cancer. J Clin Oncol. 2022; 40:2408–19.
Article7. Li S, Lai H, Liu J, Liu Y, Jin L, Li Y, et al. Circulating tumor DNA predicts the response and prognosis in patients with early breast cancer receiving neoadjuvant chemotherapy. JCO Precis Oncol. 2020; 4:PO.19.00292.
Article8. ClinicalTrials.gov. A trial using ctDNA blood tests to detect cancer cells after standard treatment to trigger additional treatment in early stage triple negative breast cancer patients (c-TRAK-TN) [Internet]. Bethesda, MD: National Library of Medicine;2017. [cited 2023 Dec 3]. Available from: https://clinicaltrials.gov/ct2/show/NCT03145961.9. ClinicalTrials.gov. A prospective, phase II trial using ctDNA to initiate post-operation boost therapy after adjuvant chemotherapy in TNBC (Artemis) [Internet]. Bethesda, MD: National Library of Medicine;2017. cited 2023 Dec 3]. Available from: https://clinicaltrials.gov/ct2/show/NCT04803539.10. ClinicalTrials.gov. A prospective, phase II trial using ctDNA to initiate post-operation boost therapy after NAC in TNBC (Apollo) [Internet]. Bethesda, MD: National Library of Medicine;2020. [cited 2023 Dec 3]. Available from: https://clinicaltrials.gov/ct2/show/NCT04501523.11. Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016; 534:47–54.12. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016; 7:11479.13. Lang GT, Jiang YZ, Shi JX, Yang F, Li XG, Pei YC, et al. Characterization of the genomic landscape and actionable mutations in Chinese breast cancers by clinical sequencing. Nat Commun. 2020; 11:5679.
Article14. Kingston B, Cutts RJ, Bye H, Beaney M, Walsh-Crestani G, Hrebien S, et al. Genomic profile of advanced breast cancer in circulating tumour DNA. Nat Commun. 2021; 12:2423.
Article15. National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology version 4 [Internet]. Plymouth Meeting, PA: National Comprehensive Cancer Network;2023 [cited 2023 Dec 3]. Available from: https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf.16. Zhang JT, Liu SY, Gao W, Liu SM, Yan HH, Ji L, et al. Longitudinal undetectable molecular residual disease defines potentially cured population in localized non-dmall vell lung cancer. Cancer Discov. 2022; 12:1690–701.17. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018; 28:1747–56.
Article18. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018; 173:321–37.19. Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016; 17:31.
Article20. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–21.21. Catalogue of Somatic Mutations in Cancer. Mutational Signatures (accessed version 2, 2015) [Internet]. Cambridgeshire: Wellcome Sanger Institute;2015. [cited 2023 Dec 3]. Available from: https://cancer.sanger.ac.uk/signatures/signatures_v2/.22. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017; 2017:PO.17.00011.
Article23. Powles T, Assaf ZJ, Davarpanah N, Banchereau R, Szabados BE, Yuen KC, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021; 595:432–7.
Article24. Fakhri N, Chad MA, Lahkim M, Houari A, Dehbi H, Belmouden A, et al. Risk factors for breast cancer in women: an update review. Med Oncol. 2022; 39:197.
Article25. van Leeuwen FE, Ronckers CM. Anthracyclines and alkylating agents: new risk factors for breast cancer in childhood cancer survivors? J Clin Oncol. 2016; 34:891–4.
Article26. Olave MC, Graham RP. Mismatch repair deficiency: the what, how and why it is important. Genes Chromosomes Cancer. 2022; 61:314–21.
Article27. Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. 2017; 2017:PO.17.00073.
Article28. Sajjadi E, Venetis K, Piciotti R, Invernizzi M, Guerini-Rocco E, Haricharan S, et al. Mismatch repair-deficient hormone receptor-positive breast cancers: biology and pathological characterization. Cancer Cell Int. 2021; 21:266.
Article29. Cakan E, Gunaydin G. Activation induced cytidine deaminase: an old friend with new faces. Front Immunol. 2022; 13:965312.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Usefulness of Ultrasonography for Detection of Breast Cancer in Patients under 30 Years of Age
- Hormone Therapy for Metastatic Breast Cancer
- Imaging features of breast cancer molecular subtypes: state of the art
- Circulating Tumor Cells Detected by RT-PCR for CK-20 before Surgery Indicate Worse Prognostic Impact in Triple-Negative and HER2 Subtype Breast Cancer
- Canine as a Comparative and Translational Model for Human Mammary Tumor