Ann Lab Med.
2024 Mar;44(2):164-173. 10.3343/alm.2023.0187.
Identification of Potential Genomic Alterations Using Pan-Cancer Cell-Free DNA Next-Generation Sequencing in Patients With Gastric Cancer
- Affiliations
-
- 1Division of Biotechnology, Invites BioCore Co. Ltd., Yongin, Korea
- 2Genome Service Development, Invites Genomics Co. Ltd., Jeju, Korea
- 3Department of Laboratory Medicine, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 4Division of Medical Oncology, Department of Internal Medicine, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- KMID: 2553386
- DOI: http://doi.org/10.3343/alm.2023.0187
Abstract
- Background
Molecular cancer profiling may lead to appropriate trials for molecularly targeted therapies. Cell-free DNA (cfDNA) is a promising diagnostic and/or prognostic biomarker in gastric cancer (GC). We characterized somatic genomic alterations in cfDNA of patients with GC.
Methods
Medical records and cfDNA data of 81 patients diagnosed as having GC were reviewed. Forty-nine and 32 patients were tested using the Oncomine Pan-Cancer CellFree Assay on the Ion Torrent platform and AlphaLiquid 100 kit on the Illumina platform, respectively.
Results
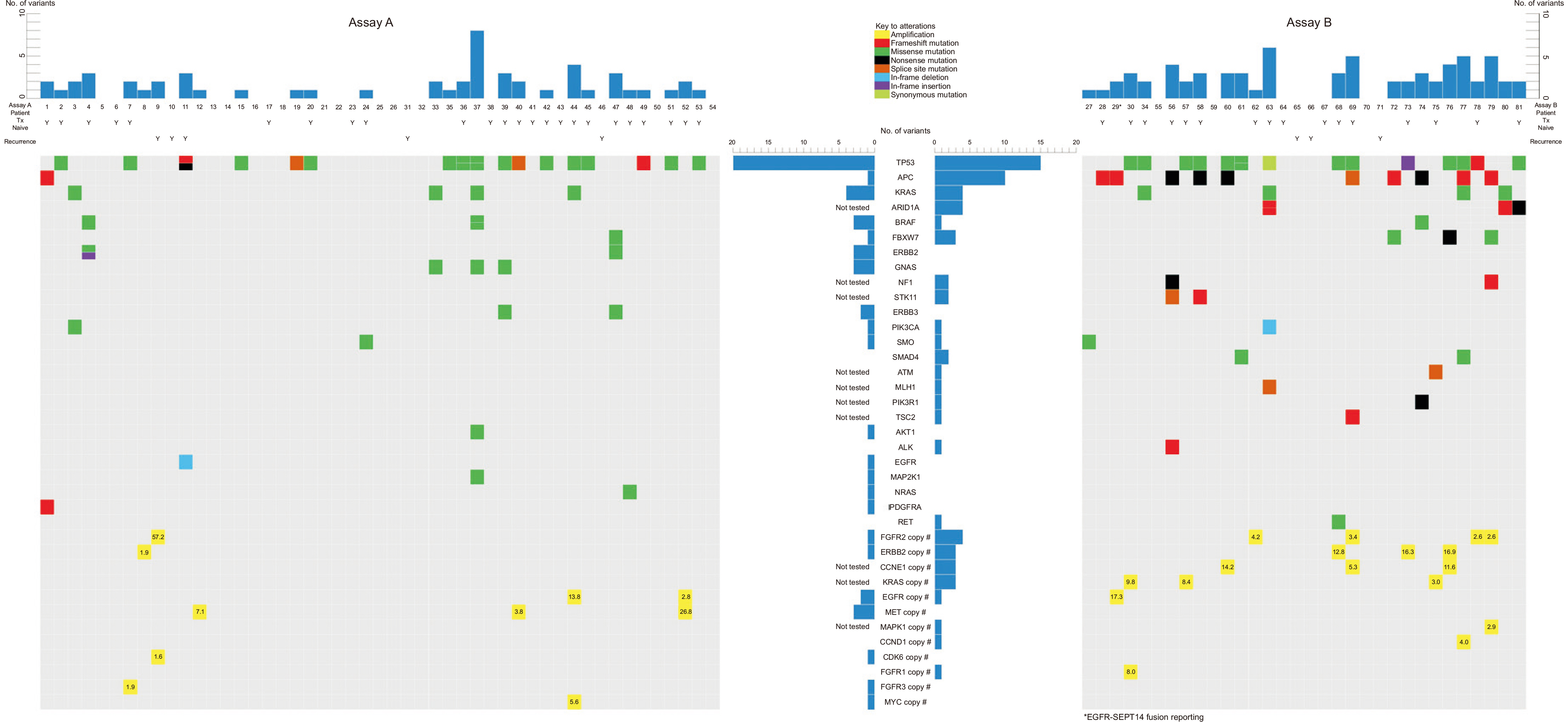
Tier I or II alterations were detected in 64.2% (52/81) of patients. Biomarkers for potential targeted therapy were detected in 55.6% of patients (45/81), and clinical trials are underway. ERBB2 amplification is actionable and was detected in 4.9% of patients (4/81). Among biomarkers showing potential for possible targeted therapy, TP53 mutation (38.3%, 35 variants in 31 patients, 31/81) and FGFR2 amplification (6.2%, 5/81) were detected the most.
Conclusions
Next-generation sequencing of cfDNA is a promising technique for the molecular profiling of GC. Evidence suggests that cfDNA analysis can provide accurate and reliable information on somatic genomic alterations in patients with GC, potentially replacing tissue biopsy as a diagnostic and prognostic tool. Through cfDNA analysis for molecular profiling, it may be possible to translate the molecular classification into therapeutic targets and predictive biomarkers, leading to personalized treatment options for patients with GC in the future.
Keyword
Figure
Cited by 1 articles
-
Next-Generation Sequencing-Based Molecular Profiling Using Cell-Free DNA: A Valuable Tool for the Diagnostic and Prognostic Evaluation of Patients With Gastric Cancer
Mi-Ae Jang
Ann Lab Med. 2024;44(2):119-121. doi: 10.3343/alm.2023.0391.
Reference
-
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; 71:209–49. DOI: 10.3322/caac.21660. PMID: 33538338.
Article2. Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. 2015; Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 21:449–56. DOI: 10.1038/nm.3850. PMID: 25894828.
Article3. Cancer Genome Atlas Research Network. 2014; Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 513:202–9. DOI: 10.1038/nature13480. PMID: 25079317. PMCID: PMC4170219.4. Kim Y, Cho MY, Kim J, Kim SN, Oh SC, Lee KA. 2017; Profiling cancer-associated genetic alterations and molecular classification of cancer in Korean gastric cancer patients. Oncotarget. 8:69888–905. DOI: 10.18632/oncotarget.19435. PMID: 29050249. PMCID: PMC5642524.
Article5. National Cancer Institute. SEER*Explorer: an interactive website for SEER cancer statistics [Internet]. Surveillance Research Program. https://seer.cancer.gov/statistics-network/explorer/. Updated on Jun 2023.6. National Cancer Information Center. 5-year cancer relative survival rates by SEER summary stage. https://www.cancer.go.kr/lay1/S1T648C652/contents.do. Updated on Jan 2023.7. National Comprehensive Cancer Network Guidelines for Gastric Cancer (Version 1.2023). https://www.nccn.org/professionals/physician_gls/pdf/gastric.pdf. Updated on Mar 2023.8. De Mattos-Arruda L, Siravegna G. 2021; How to use liquid biopsies to treat patients with cancer. ESMO Open. 6:100060. DOI: 10.1016/j.esmoop.2021.100060. PMID: 33647598. PMCID: PMC7921754.
Article9. Martínez-Saéz O, Chic N, Pascual T, Adamo B, Vidal M, González-Farré B, et al. 2020; Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 22:45. DOI: 10.1186/s13058-020-01284-9. PMID: 32404150. PMCID: PMC7222307.10. Kwapisz D. 2017; The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med. 5:46. DOI: 10.21037/atm.2017.01.32. PMID: 28251125. PMCID: PMC5326656.
Article11. Grávalos C, Gómez-Martín C, Rivera F, Alés I, Queralt B, Márquez A, et al. 2011; Phase II study of trastuzumab and cisplatin as first-line therapy in patients with HER2-positive advanced gastric or gastroesophageal junction cancer. Clin Transl Oncol. 13:179–84. DOI: 10.1007/s12094-011-0637-6. PMID: 21421462.
Article12. Lee J, Kim ST, Kim K, Lee H, Kozarewa I, Mortimer PGS, et al. 2019; Tumor genomic profiling guides patients with metastatic gastric cancer to targeted treatment: the VIKTORY umbrella trial. Cancer Discov. 9:1388–405. DOI: 10.1158/2159-8290.CD-19-0442. PMID: 31315834.
Article13. Hur JY, Chao J, Kim K, Kim ST, Kim KM, Klempner SJ, et al. 2020; High-level FGFR2 amplification is associated with poor prognosis and Lower response to chemotherapy in gastric cancers. Pathol Res Pract. 216:152878. DOI: 10.1016/j.prp.2020.152878. PMID: 32089408.
Article14. Merz V, Zecchetto C, Simionato F, Cavaliere A, Casalino S, Pavarana M, et al. 2020; A phase II trial of the FGFR inhibitor pemigatinib in patients with metastatic esophageal-gastric junction/gastric cancer trastuzumab resistant: the FiGhTeR trial. Ther Adv Med Oncol. 12:1758835920937889. DOI: 10.1177/1758835920937889. PMID: 32684989. PMCID: PMC7346700.
Article15. Nakamura Y, Kawazoe A, Lordick F, Janjigian YY, Shitara K. 2021; Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: an emerging paradigm. Nat Rev Clin Oncol. 18:473–87. DOI: 10.1038/s41571-021-00492-2. PMID: 33790428.
Article16. Shin S, Woo HI, Kim JW, Kim Y, Lee KA. 2022; Clinical practice guidelines for pre-analytical procedures of plasma epidermal growth factor receptor variant testing. Ann Lab Med. 42:141–9. DOI: 10.3343/alm.2022.42.2.141. PMID: 34635607. PMCID: PMC8548242.
Article17. Li H, Durbin R. 2009; Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–60. DOI: 10.1093/bioinformatics/btp324. PMID: 19451168. PMCID: PMC2705234.
Article18. Chen S, Liu M, Huang T, Liao W, Xu M, Gu J. 2018; GeneFuse: detection and visualization of target gene fusions from DNA sequencing data. Int J Biol Sci. 14:843–8. DOI: 10.7150/ijbs.24626. PMID: 29989075. PMCID: PMC6036752.
Article19. Gawroński AR, Lin YY, McConeghy B, LeBihan S, Asghari H, Koçkan C, et al. 2019; Structural variation and fusion detection using targeted sequencing data from circulating cell free DNA. Nucleic Acids Res. 47:e38. DOI: 10.1093/nar/gkz067. PMID: 30759232. PMCID: PMC6468241.
Article20. Deveson IW, Gong B, Lai K, LoCoco JS, Richmond TA, Schageman J, et al. 2021; Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat Biotechnol. 39:1115–28. DOI: 10.1038/s41587-021-00857-z. PMID: 33846644. PMCID: PMC8434938.
Article21. Analytical validation of the Oncomine Pan-Cancer Cell-Free Assay. White Paper. ThermoFisher Scientific. COL33100 0619, 2019. https://assets.thermofisher.com/TFS-Assets/CSD/Reference-Materials/oncomine-pan-cancer-assay-white-paper.pdf.22. So MK, Park JH, Kim JW, Jang JH. 2021; Analytical validation of a pan-cancer panel for cell-free assay for the detection of EGFR mutations. Diagnostics (Basel). 11:1022. DOI: 10.3390/diagnostics11061022. PMID: 34199654. PMCID: PMC8227964.
Article23. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. 2017; Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 19:4–23. DOI: 10.1016/j.jmoldx.2016.10.002. PMID: 27993330. PMCID: PMC5707196.
Article24. Kim B, Kim Y, Park I, Cho JY, Lee KA. 2021; Detection of EGFR-SEPT14 fusion in cell-free DNA of a patient with advanced gastric cancer: a case report. World J Clin Cases. 9:2884–9. DOI: 10.12998/wjcc.v9.i12.2884. PMID: 33969073. PMCID: PMC8058666.25. Kim S, Lim Y, Kang JK, Kim HP, Roh H, Kim SY, et al. 2022; Dynamic changes in longitudinal circulating tumour DNA profile during metastatic colorectal cancer treatment. Br J Cancer. 127:898–907. DOI: 10.1038/s41416-022-01837-z. PMID: 35643791. PMCID: PMC9427785.
Article26. Li Y, Lea K, Kshatriya P, Cao R, Gu J, Schageman J, et al. 2018; PO-086 an efficient Ion Torrent™ next generation sequencing workflow for liquid biopsy research to assess cell-free total nucleic acid. ESMO Open. 3:A260. DOI: 10.1136/esmoopen-2018-EACR25.614.
Article27. Shah M, Takayasu T, Zorofchian Moghadamtousi S, Arevalo O, Chen M, Lan C, et al. 2021; Evaluation of the Oncomine Pan-Cancer Cell-Free Assay for analyzing circulating tumor DNA in the cerebrospinal fluid in patients with central nervous system malignancies. J Mol Diagn. 23:171–80. DOI: 10.1016/j.jmoldx.2020.10.013. PMID: 33531134. PMCID: PMC7874332.
Article28. Samorodnitsky E, Jewell BM, Hagopian R, Miya J, Wing MR, Lyon E, et al. 2015; Evaluation of hybridization capture versus amplicon-based methods for whole-exome sequencing. Hum Mutat. 36:903–14. DOI: 10.1002/humu.22825. PMID: 26110913. PMCID: PMC4832303.
Article29. He Y, Sun MM, Zhang GG, Yang J, Chen KS, Xu WW, et al. 2021; Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct Target Ther. 6:425. DOI: 10.1038/s41392-021-00828-5. PMID: 34916492. PMCID: PMC8677728.
Article30. Levantini E, Maroni G, Del Re M, Tenen DG. 2022; EGFR signaling pathway as therapeutic target in human cancers. Semin Cancer Biol. 85:253–75. DOI: 10.1016/j.semcancer.2022.04.002. PMID: 35427766.
Article31. Degirmenci U, Wang M, Hu J. Targeting aberrant RAS/RAF/MEK/ERK signaling for cancer therapy. Cells. 2020; 9:DOI: 10.3390/cells9010198. PMID: 31941155. PMCID: PMC7017232.
Article32. Min A, Kim JE, Kim YJ, Lim JM, Kim S, Kim JW, et al. 2018; Cyclin E overexpression confers resistance to the CDK4/6 specific inhibitor palbociclib in gastric cancer cells. Cancer Lett. 430:123–32. DOI: 10.1016/j.canlet.2018.04.037. PMID: 29729292.
Article33. Pang W, Li Y, Guo W, Shen H. 2020; Cyclin E: a potential treatment target to reverse cancer chemoresistance by regulating the cell cycle. Am J Transl Res. 12:5170–87.34. Gu Y, Zhang P, Wang J, Lin C, Liu H, Li H, et al. 2023; Somatic ARID1A mutation stratifies patients with gastric cancer to PD-1 blockade and adjuvant chemotherapy. Cancer Immunol Immunother. 72:1199–208. DOI: 10.1007/s00262-022-03326-x. PMID: 36369379. PMCID: PMC10110689.
Article35. Qadir J, Majid S, Khan MS, Rashid F, Wani MD, Bhat SA. 2021; Implication of ARID1A undercurrents and PDL1, TP53 overexpression in advanced gastric cancer. Pathol Oncol Res. 27:1609826. DOI: 10.3389/pore.2021.1609826. PMID: 34924820. PMCID: PMC8677663.
Article36. Fang DC, Luo YH, Yang SM, Li XA, Ling XL, Fang L. 2002; Mutation analysis of APC gene in gastric cancer with microsatellite instability. World J Gastroenterol. 8:787–91. DOI: 10.3748/wjg.v8.i5.787. PMID: 12378616. PMCID: PMC4656562.
Article37. Lee JH, Choi KD, Jung HY, Baik GH, Park JK, Kim SS, et al. 2018; Seroprevalence of Helicobacter pylori in Korea: a multicenter, nationwide study conducted in 2015 and 2016. Helicobacter. 23:e12463. DOI: 10.1111/hel.12463. PMID: 29345022. PMCID: PMC5900911.
Article38. Rahman R, Asombang AW, Ibdah JA. 2014; Characteristics of gastric cancer in Asia. World J Gastroenterol. 20:4483–90. DOI: 10.3748/wjg.v20.i16.4483. PMID: 24782601. PMCID: PMC4000485.
Article39. Kim HS, Kim JH, Jang HJ. 2019; Pathologic and prognostic impacts of FGFR2 amplification in gastric cancer: a meta-analysis and systemic review. J Cancer. 10:2560–7. DOI: 10.7150/jca.29184. PMID: 31258762. PMCID: PMC6584337.40. Zheng S, Li H, Feng J, Jiang C, Lin Y, Xie Y, et al. 2022; Complete remission in leptomeningeal metastasis of NSCLC with rare EGFR-SEPT14 fusion treated with osimertinib combined with intrathecal chemotherapy with pemetrexed. Anticancer Drugs. 33:e795–8. DOI: 10.1097/CAD.0000000000001222. PMID: 34486539.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Utility of Next-Generation Sequencing for Deciphering Intratumor Heterogeneity in Prostate Cancer
- Next-Generation Sequencing-Based Molecular Profiling Using Cell-Free DNA: A Valuable Tool for the Diagnostic and Prognostic Evaluation of Patients With Gastric Cancer
- Molecular Pathology of Gastric Cancer
- Next-Generation Sequencing in Prostate Cancer
- Genomic Characteristics and the Potential Clinical Implications in Oligometastatic Non–Small Cell Lung Cancer