J Korean Med Sci.
2024 Jan;39(2):e16. 10.3346/jkms.2024.39.e16.
JAK2 Loss Arising From Tumor-SpreadThrough-Air-Spaces (STAS) Promotes Tumor Progression by Suppressing CD8+ T Cells in Lung Adenocarcinoma: A Machine Learning Approach
- Affiliations
-
- 1Department of Thoracic and Cardiovascular Surgery, Hanyang University Guri Hospital, Hanyang University College of Medicine, Guri, Korea
- 2Division of Breast Surgery, Department of Surgery, Hanyang University Guri Hospital, Hanyang University College of Medicine, Guri, Korea
- 3Department of Pathology, Uijeongbu Eulji Medical Center, Eulji University School of Medicine, Uijeongbu, Korea
- 4Department of Computer Science, Hanyang University, Seoul, Korea
- 5School of Computational Sciences, Korea Institute for Advanced Study, Seoul, Korea
- 6Department of Pathology, College of Medicine, Hanyang University, Seoul, Korea
- 7Department of Internal Medicine, Hanyang University Guri Hospital, Hanyang University College of Medicine, Guri, Korea
- 8Department of Obstetrics and Gynecology, Hanyang University Guri Hospital, Hanyang University College of Medicine, Guri, Korea
- 9Department of Pathology, Hallym University Sacred Heart Hospital, Hallym University College of Medicine, Anyang, Korea
- 10Department of Pathology, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea
- 11Department of Internal Medicine, Uijeongbu Eulji Medical Center, Eulji University School of Medicine, Uijeongbu, Korea
- KMID: 2550798
- DOI: http://doi.org/10.3346/jkms.2024.39.e16
Abstract
- Background
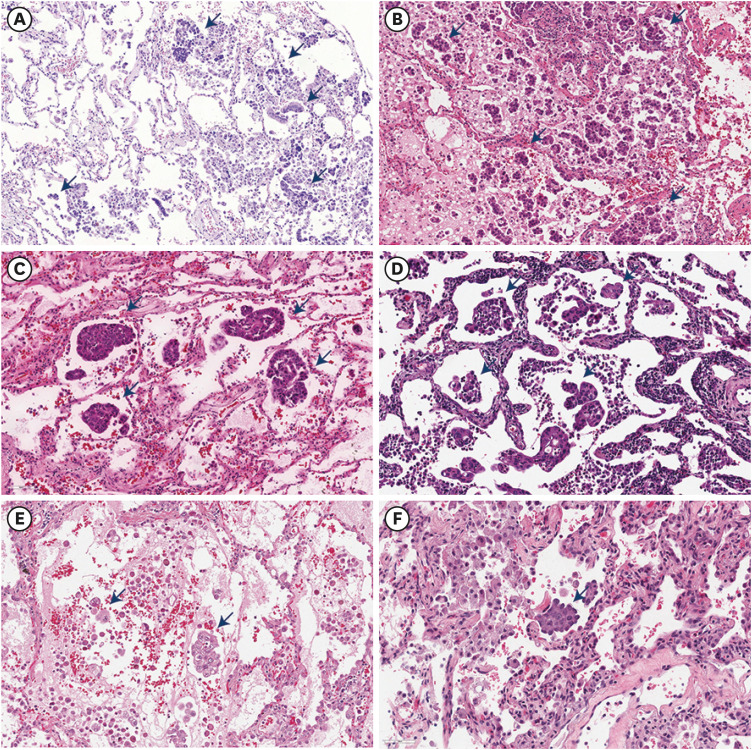
Tumor spread through air spaces (STAS) is a recently discovered risk factor for lung adenocarcinoma (LUAD). The aim of this study was to investigate specific genetic alterations and anticancer immune responses related to STAS. By using a machine learning algorithm and drug screening in lung cancer cell lines, we analyzed the effect of Janus kinase 2 (JAK2) on the survival of patients with LUAD and possible drug candidates.
Methods
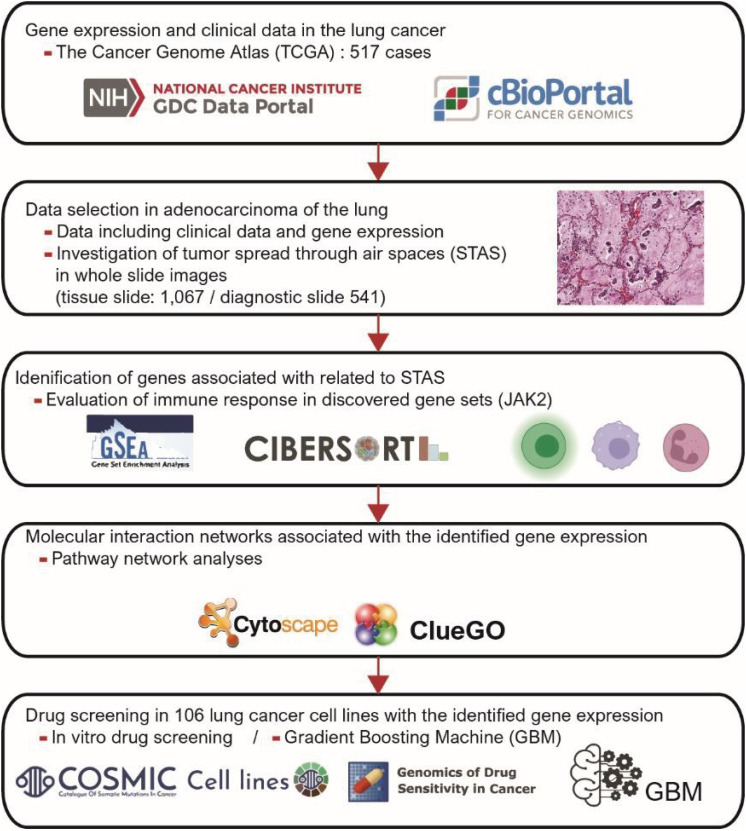
This study included 566 patients with LUAD corresponding to clinicopathological and genetic data. For analyses of LUAD, we applied gene set enrichment analysis (GSEA), in silico cytometry, pathway network analysis, in vitro drug screening, and gradient boosting machine (GBM) analysis.
Results
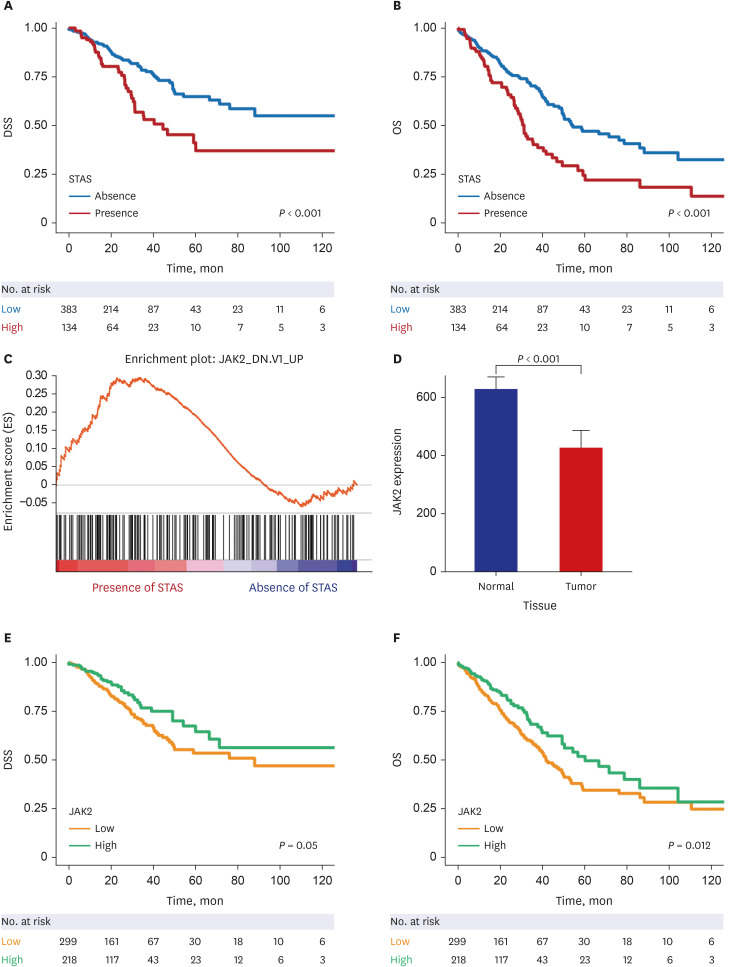
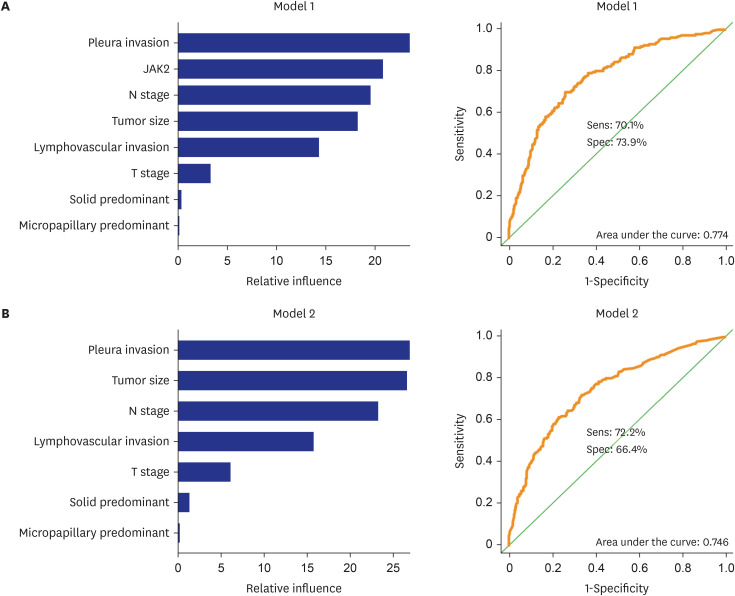
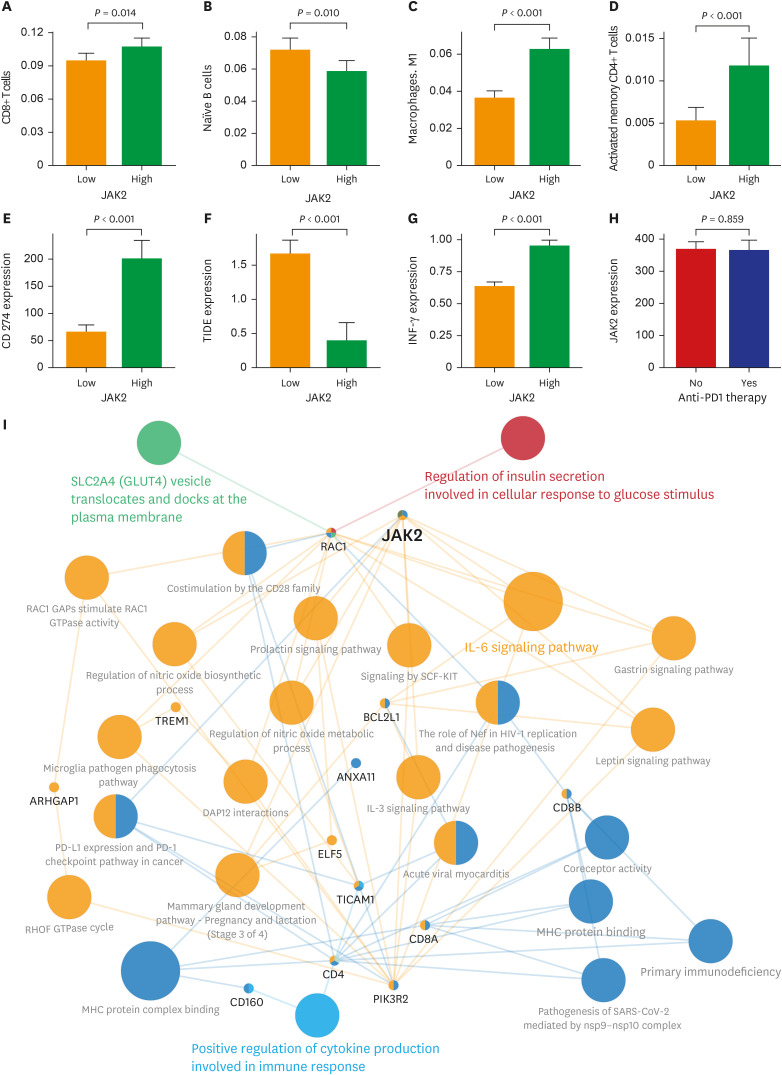
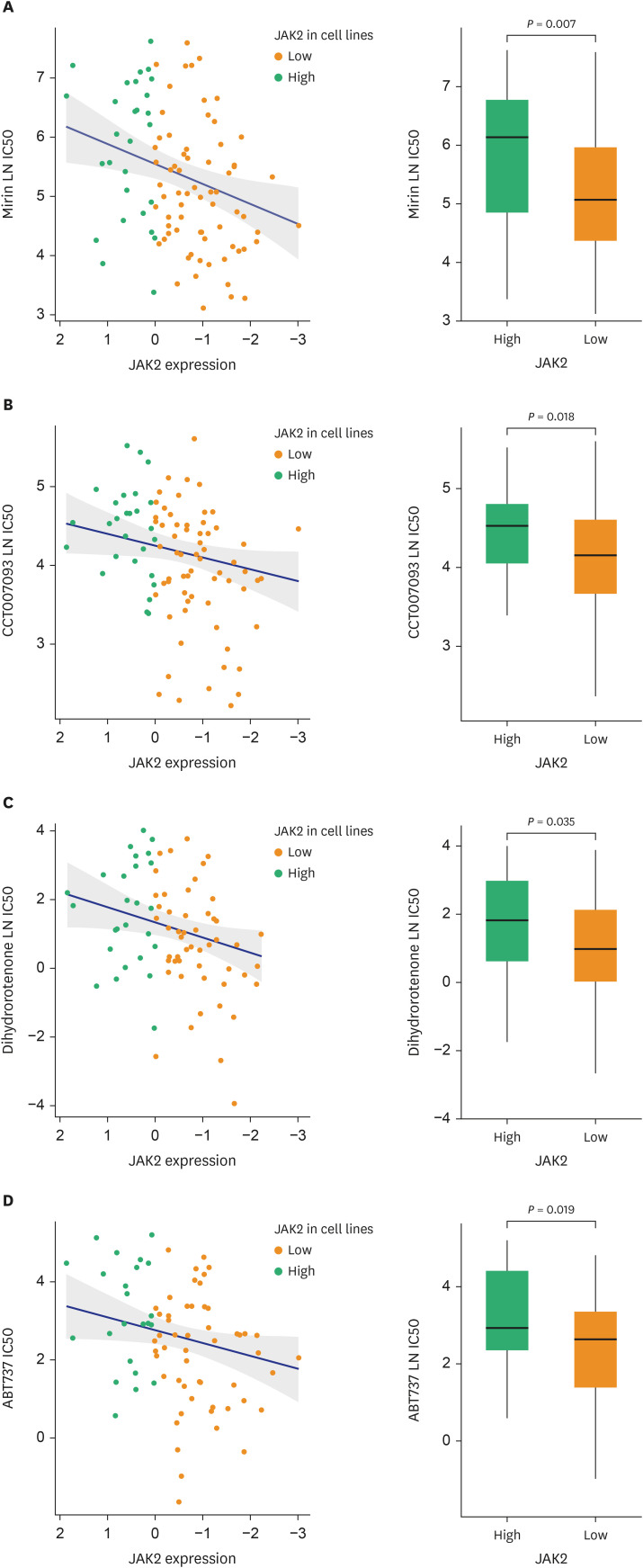
The patients with STAS had a shorter survival time than those without STAS (P < 0.001). We detected gene set-related downregulation of JAK2 associated with STAS using GSEA. Low JAK2 expression was related to poor prognosis and a low CD8+ T-cell fraction. In GBM, JAK2 showed improved survival prediction performance when it was added to other parameters (T stage, N stage, lymphovascular invasion, pleural invasion, tumor size). In drug screening, mirin, CCT007093, dihydroretenone, and ABT737 suppressed the growth of lung cancer cell lines with low JAK2 expression.
Conclusion
In LUAD, low JAK2 expression linked to the presence of STAS might serve as an unfavorable prognostic factor. A relationship between JAK2 and CD8+ T cells suggests that STAS is indirectly related to the anticancer immune response. These results may contribute to the design of future experimental research and drug development programs for LUAD with STAS.
Keyword
Figure
Reference
-
1. Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The eighth edition lung cancer stage classification. Chest. 2017; 151(1):193–203. PMID: 27780786.2. Wang C, Wu Y, Shao J, Liu D, Li W. Clinicopathological variables influencing overall survival, recurrence and post-recurrence survival in resected stage I non-small-cell lung cancer. BMC Cancer. 2020; 20(1):150. PMID: 32093621.3. Yun JK, Lee HP, Lee GD, Kim HR, Kim YH, Kim DK, et al. Recent trends in demographics, surgery, and prognosis of patients with surgically resected lung cancer in a single institution from Korea. J Korean Med Sci. 2019; 34(45):e291. PMID: 31760712.4. Ekeke CN, Mitchell C, Schuchert M, Dhupar R, Luketich JD, Okusanya OT. Early distant recurrence in patients with resected stage I lung cancer: a case series of “Blast Metastasis”. Clin Lung Cancer. 2021; 22(1):e132–e135. PMID: 33144072.5. Masago K, Seto K, Fujita S, Sasaki E, Hosoda W, Kuroda H. Long-term recurrence of completely resected NSCLC. JTO Clin Res Rep. 2020; 1(3):100076. PMID: 34589953.6. Seung SJ, Hurry M, Walton RN, Evans WK. Real-world treatment patterns and survival in stage IV non-small-cell lung cancer in Canada. Curr Oncol. 2020; 27(4):e361–e367. PMID: 32905294.7. Miller M, Hanna N. Advances in systemic therapy for non-small cell lung cancer. BMJ. 2021; 375(2363):n2363. PMID: 34753715.8. Park S. Dancing with the surgeon: neoadjuvant and adjuvant immunotherapies from the medical oncologist’s perspective. J Chest Surg. 2023; 56(2):67–74. PMID: 36864673.9. Jung JM, Hyun SJ, Kim KJ. Surgical impacts of metastatic non-small cell lung cancer to the thoracic and lumbar spine. J Korean Med Sci. 2021; 36(7):e52. PMID: 33619918.10. Reck M, Remon J, Hellmann MD. First-line immunotherapy for non-small-cell lung cancer. J Clin Oncol. 2022; 40(6):586–597. PMID: 34985920.11. Brahmer JR, Lee JS, Ciuleanu TE, Bernabe Caro R, Nishio M, Urban L, et al. Five-year survival outcomes with nivolumab plus ipilimumab versus chemotherapy as first-line treatment for metastatic non-small-cell lung cancer in CheckMate 227. J Clin Oncol. 2023; 41(6):1200–1212. PMID: 36223558.12. Isbell JM, Li BT, Gomez DR. The emerging role of local therapy in oligometastatic non-small cell lung cancer. J Thorac Cardiovasc Surg. 2022; 163(3):819–825. PMID: 34147255.13. Yoo S, Cho WC, Lee GD, Choi S, Kim HR, Kim YH, et al. Long-term surgical outcomes in oligometastatic non-small cell lung cancer: a single-center study. J Chest Surg. 2023; 56(1):25–32. PMID: 36517949.14. Jung YJ, Oh IJ, Kim Y, Jung JH, Seok M, Lee W, et al. Clinical validation of a protein biomarker panel for non-small cell lung cancer. J Korean Med Sci. 2018; 33(53):e342. PMID: 30595683.15. Kim DJ, Kim WJ, Lim M, Hong Y, Lee SJ, Hong SH, et al. Plasma CRABP2 as a novel biomarker in patients with non-small cell lung cancer. J Korean Med Sci. 2018; 33(26):e178. PMID: 29930489.16. Lee S, Kang HG, Choi JE, Lee JH, Kang HJ, Baek SA, et al. The different effect of VEGF polymorphisms on the prognosis of non-small cell lung cancer according to tumor histology. J Korean Med Sci. 2016; 31(11):1735–1741. PMID: 27709850.17. Warth A, Muley T, Kossakowski CA, Goeppert B, Schirmacher P, Dienemann H, et al. Prognostic impact of intra-alveolar tumor spread in pulmonary adenocarcinoma. Am J Surg Pathol. 2015; 39(6):793–801. PMID: 25723114.18. Kadota K, Nitadori JI, Sima CS, Ujiie H, Rizk NP, Jones DR, et al. Tumor spread through air spaces is an important pattern of invasion and impacts the frequency and location of recurrences after limited resection for small stage I lung adenocarcinomas. J Thorac Oncol. 2015; 10(5):806–814. PMID: 25629637.19. Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. Lyon, France: IARC Press;2015.20. Yanagawa N, Shiono S, Endo M, Ogata SY. Tumor spread through air spaces is a useful predictor of recurrence and prognosis in stage I lung squamous cell carcinoma, but not in stage II and III. Lung Cancer. 2018; 120:14–21. PMID: 29748009.21. Han YB, Kim H, Mino-Kenudson M, Cho S, Kwon HJ, Lee KR, et al. Tumor spread through air spaces (STAS): prognostic significance of grading in non-small cell lung cancer. Mod Pathol. 2021; 34(3):549–561. PMID: 33199839.22. Sim J, Kim H, Hyeon J, Choi Y, Han J. Anaplastic lymphoma kinase (ALK)-expressing lung adenocarcinoma with combined neuroendocrine component or neuroendocrine transformation: implications for neuroendocrine transformation and response to ALK-tyrosine kinase inhibitors. J Korean Med Sci. 2018; 33(15):e123. PMID: 29629521.23. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102(43):15545–15550. PMID: 16199517.24. Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013; 29(5):661–663. PMID: 23325622.25. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009; 25(8):1091–1093. PMID: 19237447.26. Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018; 24(10):1550–1558. PMID: 30127393.27. Yoshizawa A, Motoi N, Riely GJ, Sima CS, Gerald WL, Kris MG, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol. 2011; 24(5):653–664. PMID: 21252858.28. Jung WY, Min KW, Oh YH. Increased VEGF-A in solid type of lung adenocarcinoma reduces the patients’ survival. Sci Rep. 2021; 11(1):1321. PMID: 33446784.29. Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013; 41:D955–D961. PMID: 23180760.30. Blaauwgeers H, Flieder D, Warth A, Harms A, Monkhorst K, Witte B, et al. A prospective study of loose tissue fragments in non–small cell lung cancer resection specimens: an alternative view to “spread through air spaces”. Am J Surg Pathol. 2017; 41(9):1226–1230. PMID: 28622180.31. Thunnissen E, Blaauwgeers HJ, de Cuba EM, Yick CY, Flieder DB. Ex vivo artifacts and histopathologic pitfalls in the lung. Arch Pathol Lab Med. 2016; 140(3):212–220. PMID: 26927715.32. Ikezoe T, Kojima S, Furihata M, Yang J, Nishioka C, Takeuchi A, et al. Expression of p-JAK2 predicts clinical outcome and is a potential molecular target of acute myelogenous leukemia. Int J Cancer. 2011; 129(10):2512–2521. PMID: 21207414.33. Sonohara F, Nomoto S, Inokawa Y, Hishida M, Takano N, Kanda M, et al. High expression of Janus kinase 2 in background normal liver tissue of resected hepatocellular carcinoma is associated with worse prognosis. Oncol Rep. 2015; 33(2):767–773. PMID: 25420511.34. Song Y, Tang MY, Chen W, Wang Z, Wang SL. High JAK2 protein expression predicts a poor prognosis in patients with resectable pancreatic ductal adenocarcinoma. Dis Markers. 2020; 2020:7656031. PMID: 33029256.35. Economopoulou P, Kotoula V, Koliou GA, Papadopoulou K, Christodoulou C, Pentheroudakis G, et al. Prognostic impact of Src, CDKN1B, and JAK2 expression in metastatic breast cancer patients treated with trastuzumab. Transl Oncol. 2019; 12(5):739–748. PMID: 30877976.36. Ikeda S, Okamoto T, Okano S, Umemoto Y, Tagawa T, Morodomi Y, et al. PD-L1 is upregulated by simultaneous amplification of the PD-L1 and JAK2 genes in non-small cell lung cancer. J Thorac Oncol. 2016; 11(1):62–71. PMID: 26762740.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Poor Prognosis of Grade 2 Spread Through Air Spaces in Neuroendocrine Tumors
- Computed Tomography Radiomics for Preoperative Prediction of Spread Through Air Spaces in the Early Stage of Surgically Resected Lung Adenocarcinomas
- Morphological Features of the Most Advanced Intra-Tumor Component in Multistep Progression
- Proteomics analysis of tumor-infiltrated T cell reveals CD127+ and KLRG1+ memory CD8+ T cells control immunotherapy efficacy in hepatocellular carcinoma
- Progressive Increase of Regulatory T Cells and Decrease of CD8+ T Cells and CD8+ T Cells/Regulatory T Cells Ratio during Colorectal Cancer Development