Cancer Res Treat.
2024 Jan;56(1):149-161. 10.4143/crt.2023.800.
Next-Generation Sequencing in Breast Cancer Patients: Real-World Data for Precision Medicine
- Affiliations
-
- 1Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 2Department of Digital Health, Samsung Advanced Institute of Health Science and Technology, Sungkyunkwan University School of Medicine, Seoul, Korea
- 3Department of Clinical Genomic Center, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- KMID: 2550332
- DOI: http://doi.org/10.4143/crt.2023.800
Abstract
- Purpose
Breast cancer is one of the most common causes of cancer-related death in females. Numerous drug-targetable biomarkers and predictive biomarkers have been developed. Some researchers have expressed doubts about the need for next-generation sequencing (NGS) studies in daily practice. This study analyzed the results of NGS studies on breast cancer at a single institute and evaluated the real-world applications of NGS data to precision medicine for breast cancer.
Materials and Methods
We retrospectively collected the results of NGS studies and analyzed the histopathologic features and genetic profiles of patients treated for breast cancer from 2010 to 2021. Seventy cases had data from CancerSCAN, a customized panel of 375 cancer-associated genes, and 110 cases had data from TruSight Oncology 500.
Results
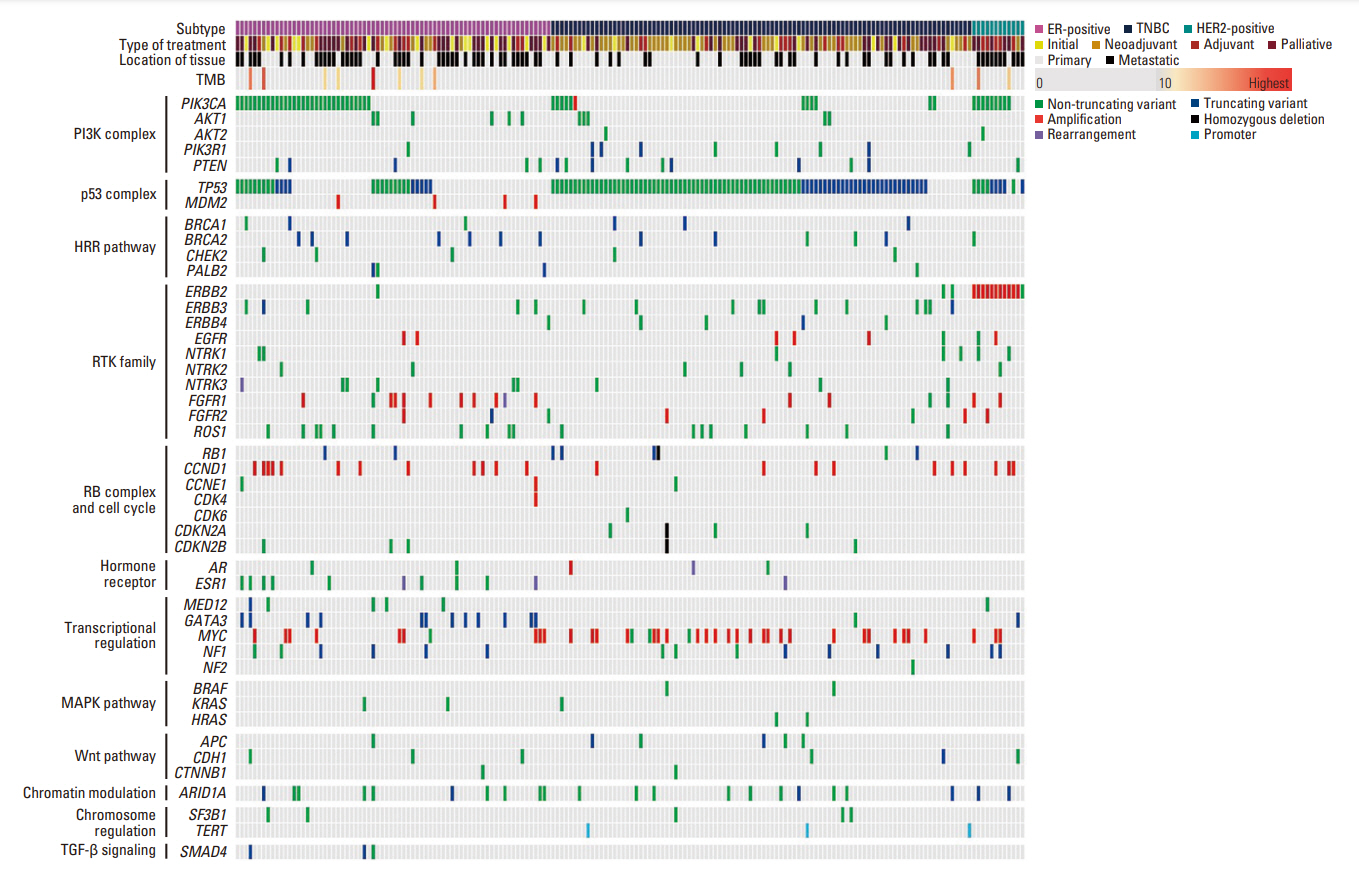
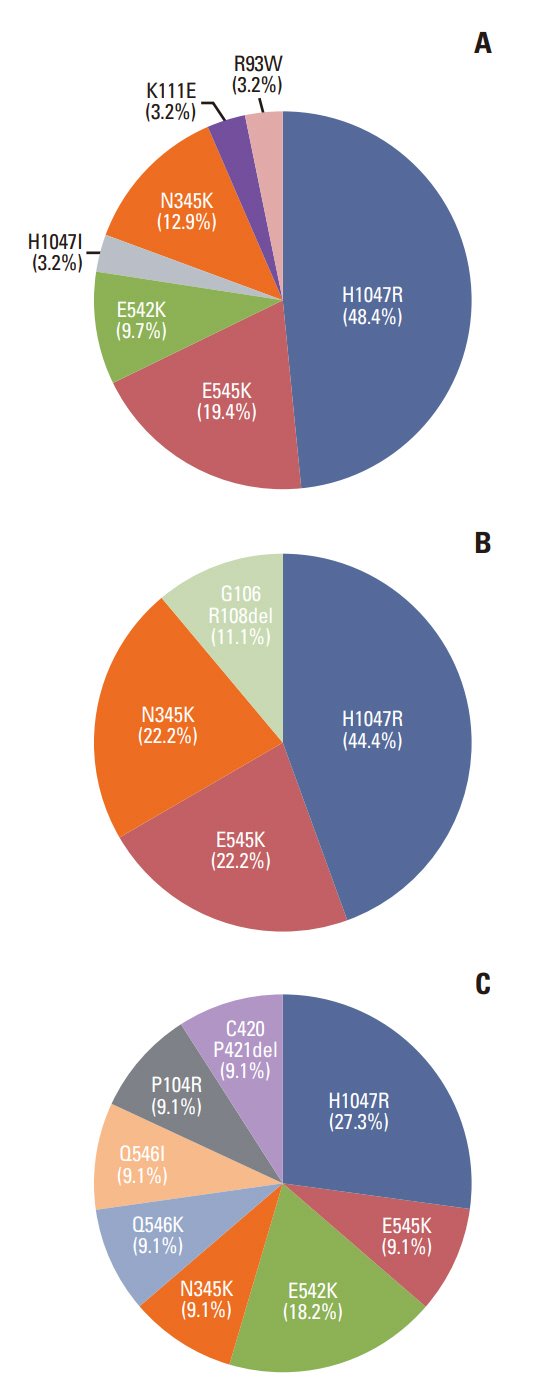
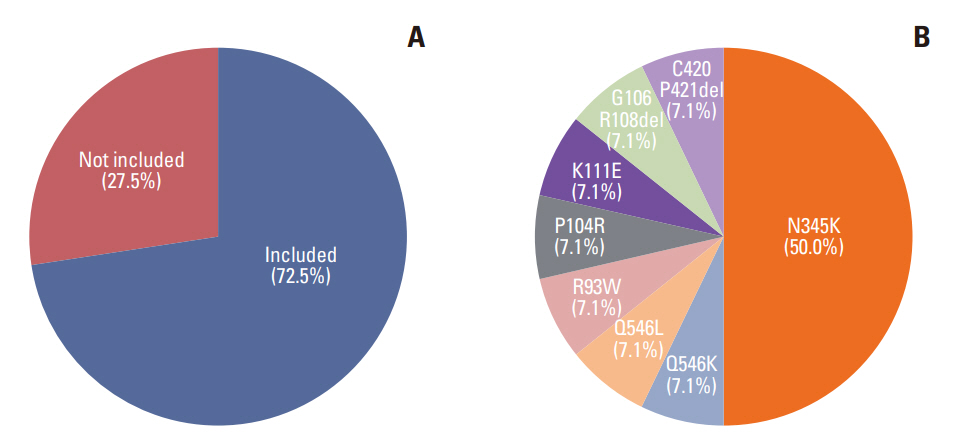
The most frequently detected single nucleotide variant was the TP53 mutation (123/180, 68.3%), followed by PIK3CA muta-tions (51/180, 28.3%). Estrogen receptor 1 (ESR1) mutation was detected in 11 patients (6.1%), of whom 10 had hormone receptor–positive, human epidermal growth factor receptor 2–negative breast cancer, and two had no history of prior endocrine therapy. Based on their NGS study results, 13 patients (7.2%) received target therapy. Among them, four patients had a BRCA1 or BRCA2 germline mutation, and nine patients had a PIK3CA mutation.
Conclusion
NGS can provide information about predictive biomarkers and drug-targetable biomarkers that can enable treatment and participation in clinical trials based on precision medicine. Further studies should be conducted to excavate novel drug-targetable biomarkers and develop additional target therapies.
Figure
Reference
-
References
1. Choi JE, Kim Z, Park CS, Park EH, Lee SB, Lee SK, et al. Breast cancer statistics in Korea, 2019. J Breast Cancer. 2023; 26:207–20.2. Hudis CA. Trastuzumab: mechanism of action and use in clinical practice. N Engl J Med. 2007; 357:39–51.3. Modi S, Saura C, Yamashita T, Park YH, Kim SB, Tamura K, et al. Trastuzumab deruxtecan in previously treated HER2-positive breast cancer. N Engl J Med. 2020; 382:610–21.4. Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998; 339:1609–18.5. Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015; 16:25–35.6. Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018; 379:2108–21.7. Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im SA, Yusof MM, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. 2020; 396:1817–28.8. Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A, et al. Breast cancer statistics, 2022. CA Cancer J Clin. 2022; 72:524–41.9. Shin HC, Lee HB, Yoo TK, Lee ES, Kim RN, Park B, et al. Detection of germline mutations in breast cancer patients with clinical features of hereditary cancer syndrome using a multi-gene panel test. Cancer Res Treat. 2020; 52:697–713.10. Han SA, Park SK, Ahn SH, Lee MH, Noh DY, Kim LS, et al. The Korean Hereditary Breast Cancer (KOHBRA) study: protocols and interim report. Clin Oncol (R Coll Radiol). 2011; 23:434–41.11. Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018; 379:111–21.12. Piccart M, van ‘t Veer LJ, Poncet C, Lopes Cardozo JM, Delaloge S, Pierga JY, et al. 70-gene signature as an aid for treatment decisions in early breast cancer: updated results of the phase 3 randomised MINDACT trial with an exploratory analysis by age. Lancet Oncol. 2021; 22:476–88.13. Li SY, Rong M, Grieu F, Iacopetta B. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat. 2006; 96:91–5.14. Wang T, Xu Y, Sheng S, Yuan H, Ouyang T, Li J, et al. HER2 somatic mutations are associated with poor survival in HER2-negative breast cancers. Cancer Sci. 2017; 108:671–7.15. Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013; 45:1439–45.16. Zheng ZY, Anurag M, Lei JT, Cao J, Singh P, Peng J, et al. Neurofibromin is an estrogen receptor-alpha transcriptional co-repressor in breast cancer. Cancer Cell. 2020; 37:387–402.17. Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020; 31:1491–505.18. Amin MB, Edge SB, Greene FL, Byrd DR, Brookland RK, Washington MK, et al. AJCC cancer staging manual. 8th ed. New York, NY: Springer;2017. p. 589–628.19. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016; 7:11479.20. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012; 486:346–52.21. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70.22. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–4.23. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013; 6:pl1.24. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017; 2017:PO.17.00011.25. Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019; 380:1929–40.26. Dogruluk T, Tsang YH, Espitia M, Chen F, Chen T, Chong Z, et al. Identification of variant-specific functions of PIK3CA by rapid phenotyping of rare mutations. Cancer Res. 2015; 75:5341–54.27. Ludwig KK, Neuner J, Butler A, Geurts JL, Kong AL. Risk reduction and survival benefit of prophylactic surgery in BRCA mutation carriers, a systematic review. Am J Surg. 2016; 212:660–9.28. Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations: a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015; 12:573–83.29. Jeselsohn R, Yelensky R, Buchwalter G, Frampton G, Meric-Bernstam F, Gonzalez-Angulo AM, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014; 20:1757–67.30. Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013; 45:1446–51.31. Bidard FC, Kaklamani VG, Neven P, Streich G, Montero AJ, Forget F, et al. Elacestrant (oral selective estrogen receptor degrader) versus standard endocrine therapy for estrogen receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: results from the randomized phase III EMERALD trial. J Clin Oncol. 2022; 40:3246–56.32. Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020; 21:1353–65.33. Winer EP, Lipatov O, Im SA, Goncalves A, Munoz-Couselo E, Lee KS, et al. Association of tumor mutational burden (TMB) and clinical outcomes with pembrolizumab (pembro) versus chemotherapy (chemo) in patients with metastatic triple-negative breast cancer (mTNBC) from KEYNOTE-119. J Clin Oncol. 2020; 38(15 Suppl):1013.34. Barroso-Sousa R, Jain E, Cohen O, Kim D, Buendia-Buendia J, Winer E, et al. Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann Oncol. 2020; 31:387–94.35. McNulty SN, Parikh BA, Duncavage EJ, Heusel JW, Pfeifer JD. Optimization of population frequency cutoffs for filtering common germline polymorphisms from tumor-only next-generation sequencing data. J Mol Diagn. 2019; 21:903–12.36. Suh KJ, Kim SH, Kim YJ, Shin H, Kang E, Kim EK, et al. Clinical application of next-generation sequencing in patients with breast cancer: real-world data. J Breast Cancer. 2022; 25:366–78.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Application of Next-Generation Sequencing in Patients With Breast Cancer: Real-World Data
- Ultra-rare Disease and Genomics-Driven Precision Medicine
- Application of precision medicine based on next-generation sequencing and immunohistochemistry in ovarian cancer: a real-world experience
- Applying Precision Medicine in Clinical Practice
- The Cancer Precision Medicine Diagnosis and Treatment (K-MASTER) Enterprise