Ann Lab Med.
2024 Jan;44(1):47-55. 10.3343/alm.2024.44.1.47.
Comparative Genomic Analysis of Staphylococcal Cassette Chromosome mec Type V Staphylococcus aureus Strains and Estimation of the Emergence of SCCmec V Clinical Isolates in Korea
- Affiliations
-
- 1Laboratory of Infectious Diseases, Graduate School of Infection Control Sciences & Ōmura Satoshi Memorial Institute, Kitasato University, Tokyo, Japan
- 2Bioresources Collection and Research Division, Bioresources Collection and Bioinformation Department, Nakdonggang National Institute of Biological Resources (NNIBR), Sangju, Korea
- 3Department of Laboratory Medicine, Hallym University College of Medicine, Seoul, Korea
- KMID: 2550222
- DOI: http://doi.org/10.3343/alm.2024.44.1.47
Abstract
- Background
Staphylococcal cassette chromosome mec type V (SCCmec V) methicillin-resistant Staphylococcus aureus (MRSA) has been recovered from patients and livestock. Using comparative genomic analyses, we evaluated the phylogenetic emergence of SCCmec V after transmission from overseas donor strains to Korean recipient strains.
Methods
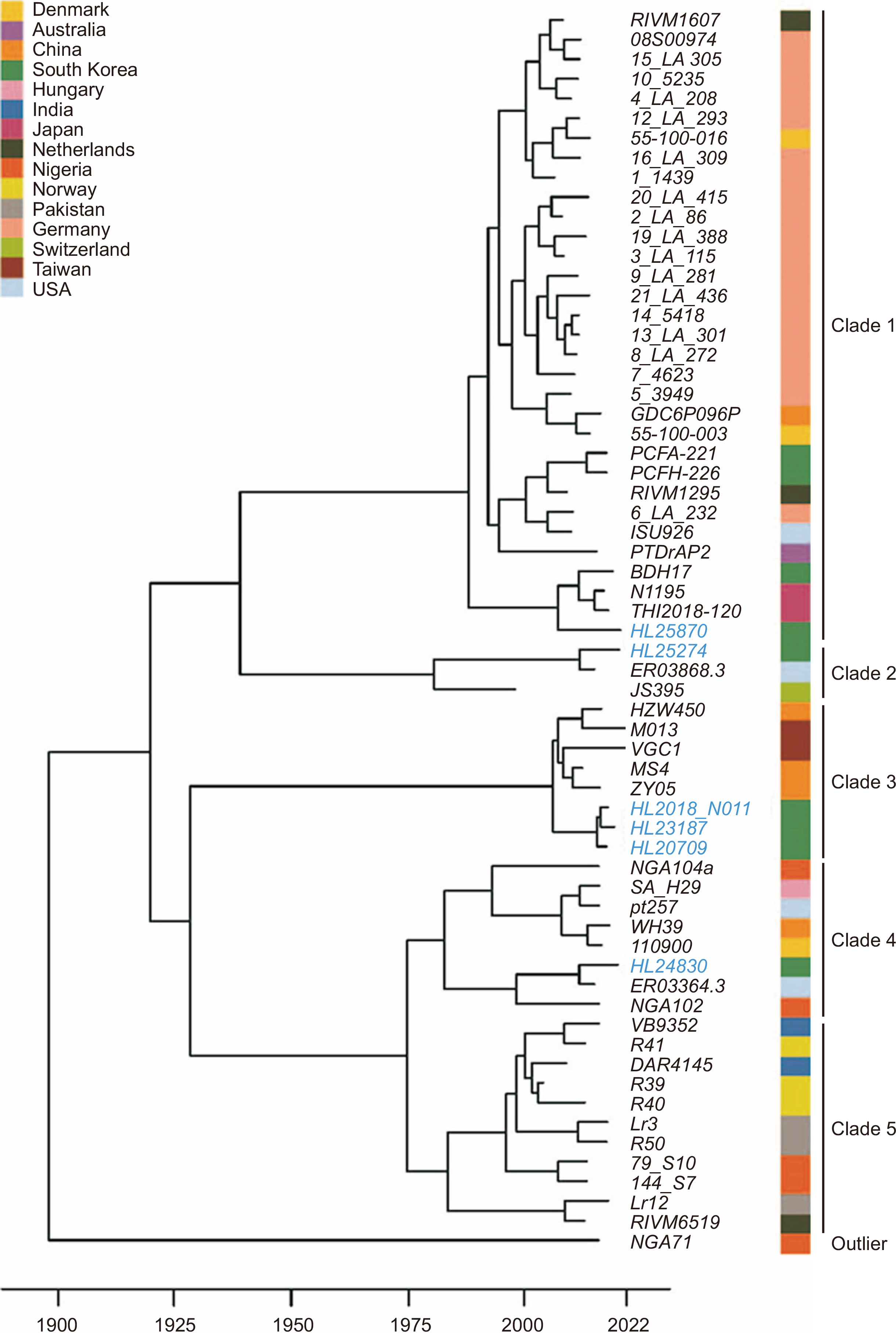
Sixty-three complete MRSA SCCmec V genomes (including six Korean clinical isolates) were used to construct a phylogenetic tree. Single-nucleotide polymorphisms were identified using Snippy, and a maximum-likelihood-based phylogenetic tree was constructed using RAxML. The possible emergence of the most common ancestor was estimated using BactDating. To estimate mecA horizontal gene transfer (HGT) events, Rangerdtl was applied to 818 SCCmec V strains using publicly available whole-genome data.
Results
The phylogenetic tree showed five major clades. German strains formed a major clade; their possible origin was traced to the 1980s. The emergence of Korean SCCmec V clinical isolates was traced to 2000–2010. mecA HGT events in Staphylococcus spp. were identified in seven strains. P7 (Hong Kong outbreak strain) served as the donor strain for two Korean sequence type (ST) 59 strains, whereas the other five recipient strains emerged from different SCCmec V donors.
Conclusions
Most Korean SCCmec V strains may have emerged during 2000–2010. A unique MRSA SCCmec V strain, ST72 (a Korean common type of community-associated MRSA), was also identified. The genomic dynamics of this clone with a zoonotic background should be monitored to accurately understand MRSA evolution.
Keyword
Figure
Reference
-
1. Park SH. 1970; Antibiotic susceptibility of pathogenic microorganisms isolated in 1969. J Korean Med Assoc. 13:337–46.2. Chong Y, Lee K. 2000; Present situation of antimicrobial resistance in Korea. J Infect Chemother. 6:189–95. DOI: 10.1007/s101560070001. PMID: 11810564.
Article3. Kim JS, Kim HS, Song W, Cho HC, Lee KM, Kim EC. 2007; Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates with toxic shock syndrome toxin and staphylococcal enterotoxin C genes. Korean J Lab Med. 27:118–23. DOI: 10.3343/kjlm.2007.27.2.118. PMID: 18094562.4. Lee SS, Kim YJ, Chung DR, Jung KS, Kim JS. 2010; Invasive infection caused by a community-associated methicillin-resistant Staphylococcus aureus strain not carrying Panton-Valentine leukocidin in South Korea. J Clin Microbiol. 48:311–3. DOI: 10.1128/JCM.00297-09. PMID: 19889903. PMCID: PMC2812302.
Article5. Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. 2003; Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 36:53–9. DOI: 10.1086/345476. PMID: 12491202.
Article6. Park JM, Kwon M, Hong KH, Lee H, Yong D. 2023; European Committee on Antimicrobial Susceptibility Testing-recommended rapid antimicrobial susceptibility testing of Escherichia coli, Klebsiella pneumoniae, and Staphylococcus aureus from positive blood culture bottles. Ann Lab Med. 43:443–50. DOI: 10.3343/alm.2023.43.5.443. PMID: 37080745. PMCID: PMC10151279.
Article7. Katayama Y, Ito T, Hiramatsu K. 2000; A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 44:1549–55. DOI: 10.1128/AAC.44.6.1549-1555.2000. PMID: 10817707. PMCID: PMC89911.
Article8. Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. 2002; The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A. 99:7687–92. DOI: 10.1073/pnas.122108599. PMID: 12032344. PMCID: PMC124322.9. Oliveira DC, Tomasz A, de Lencastre H. 2002; Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect Dis. 2:180–9. DOI: 10.1016/S1473-3099(02)00227-X. PMID: 11944188.10. Oliveira DC, de Lencastre H. 2002; Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 46:2155–61. DOI: 10.1128/AAC.46.7.2155-2161.2002. PMID: 12069968. PMCID: PMC127318.
Article11. Kim JS, Song W, Kim HS, Cho HC, Lee KM, Choi MS, et al. 2006; Association between the methicillin resistance of clinical isolates of Staphylococcus aureus, their staphylococcal cassette chromosome mec (SCCmec) subtype classification, and their toxin gene profiles. Diagn Microbiol Infect Dis. 56:289–95. DOI: 10.1016/j.diagmicrobio.2006.05.003. PMID: 16854552.12. Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. 2004; Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother. 48:2637–51. DOI: 10.1128/AAC.48.7.2637-2651.2004. PMID: 15215121. PMCID: PMC434217.
Article13. Lade H, Chung SH, Lee Y, Joo HS, Kim JS. 2022; Genotypes of Staphylococcus aureus clinical isolates are associated with phenol-soluble modulin (PSM) production. Toxins (Basel). 14:556. DOI: 10.3390/toxins14080556. PMID: 36006218. PMCID: PMC9412541.
Article14. Aires-de-Sousa M. 2017; Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin Microbiol Infect. 23:373–80. DOI: 10.1016/j.cmi.2016.11.002. PMID: 27851997.15. Wang Y, Zhang P, Wu J, Chen S, Jin Y, Long J, et al. Transmission of livestock-associated methicillin-resistant Staphylococcus aureus between animals, environment, and humans in the farm. Environ Sci Pollut Res Int. doi: 10.1007/s11356-023-28532-7. Epub ahead of print. DOI: 10.1007/s11356-023-28532-7. PMID: 37418185.16. Back SH, Eom HS, Lee HH, Lee GY, Park KT, Yang SJ. 2020; Livestock-associated methicillin-resistant Staphylococcus aureus in Korea: antimicrobial resistance and molecular characteristics of LA-MRSA strains isolated from pigs, pig farmers, and farm environment. J Vet Sci. 21:e2. DOI: 10.4142/jvs.2020.21.e2. PMID: 31940681. PMCID: PMC7000904.
Article17. Centers for Disease Control and Prevention. One Health. https://www.cdc.gov/onehealth/index.html. Updated on July 2023.18. Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, et al. 2003; Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 41:5442–8. DOI: 10.1128/JCM.41.12.5442-5448.2003. PMID: 14662923. PMCID: PMC309029.
Article19. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, et al. 2007; Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 51:264–74. DOI: 10.1128/AAC.00165-06. PMID: 17043114. PMCID: PMC1797693.
Article20. Kim JS, Sakaguchi S, Fukushima Y, Yoshida H, Nakano T, Takahashi T. 2020; Complete genome sequences of four Streptococcus canis strains isolated from dogs in South Korea. Microbiol Resour Announc. 9:e00818–20. DOI: 10.1128/MRA.00818-20. PMID: 32817161. PMCID: PMC7427199.21. Bolger AM, Lohse M, Usadel B. 2014; Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–20. DOI: 10.1093/bioinformatics/btu170. PMID: 24695404. PMCID: PMC4103590.
Article22. Grant JR, Arantes AS, Stothard P. 2012; Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genomics. 13:202. DOI: 10.1186/1471-2164-13-202. PMID: 22621371. PMCID: PMC3469350.
Article23. Kim JM, Fukushima Y, Yoshida H, Kim JS, Takahashi T. 2022; Comparative genomic features of Streptococcus canis based on pan-genome orthologous group analysis according to sequence type. Jpn J Infect Dis. 75:269–76. DOI: 10.7883/yoken.JJID.2021.533. PMID: 34588372.
Article24. Yoshida H, Kim JM, Maeda T, Goto M, Tsuyuki Y, Shibata S, et al. 2023; Virulence-associated genome sequences of Pasteurella canis and unique toxin gene prevalence of P. canis and Pasteurella multocida isolated from humans and companion animals. Ann Lab Med. 43:263–72. DOI: 10.3343/alm.2023.43.3.263. PMID: 36544338. PMCID: PMC9791007.
Article25. Thodberg K, Videbech PB, Hansen TGB, Pedersen AB, Christensen JW. 2021; Dog visits in nursing homes - increase complexity or keep it simple? A randomised controlled study. PLoS One. 16:e0251571. DOI: 10.1371/journal.pone.0251571. PMID: 34038451. PMCID: PMC8153477.
Article26. Moon BY, Youn JH, Shin S, Hwang SY, Park YH. 2012; Genetic and phenotypic characterization of methicillin-resistant staphylococci isolated from veterinary hospitals in South Korea. J Vet Diagn Invest. 24:489–98. DOI: 10.1177/1040638712440985. PMID: 22529115.
Article27. Kang JH, Hwang CY. 2021; One health approach to genetic relatedness in SCCmec between methicillin-resistant Staphylococcus isolates from companion dogs with pyoderma and their owners. Vet Microbiol. 253:108957. DOI: 10.1016/j.vetmic.2020.108957. PMID: 33385887.28. Cheng VCC, Wong SC, Cao H, Chen JHK, So SYC, Wong SCY, et al. 2019; Whole-genome sequencing data-based modeling for the investigation of an outbreak of community-associated methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit in Hong Kong. Eur J Clin Microbiol Infect Dis. 38:563–73. DOI: 10.1007/s10096-018-03458-y. PMID: 30680562.
Article29. Chlebowicz MA, Mašlaňová I, Kuntová L, Grundmann H, Pantůček R, Doškař J, et al. 2014; The staphylococcal cassette chromosome mec type V from Staphylococcus aureus ST398 is packaged into bacteriophage capsids. Int J Med Microbiol. 304:764–74. DOI: 10.1016/j.ijmm.2014.05.010. PMID: 24951306.30. Giovanni N, Elisa S, Marta C, Rosa F, Loredana C, Alessandra B, et al. 2020; Occurrence and characteristics of methicillin-resistant Staphylococcus aureus (MRSA) in buffalo bulk tank milk and the farm workers in Italy. Food Microbiol. 91:103509. DOI: 10.1016/j.fm.2020.103509. PMID: 32539967.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Correlation Between Staphylococcal Cassette Chromosome mec Type and Coagulase Serotype of Methicillin-Resistant Staphylococcus aureus
- Detection of Integrons and Staphylococcal Cassette Chromosome mec Types in Clinical Methicillin-resistant Coagulase Negative Staphylococci Strains
- Acquisition of methicillin resistance and progression of multiantibiotic resistance in methicillin-resistant Staphylococcus aureus
- Emergence of Panton-Valentine Leukocidin-Positive ST80 Clone of Community-Associated Methicillin-Resistant Staphylococcus aureus in Busan, Korea
- Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus Isolates with Toxic Shock Syndrome Toxin and Staphylococcal Enterotoxin C genes