Cancer Res Treat.
2023 Oct;55(4):1291-1302. 10.4143/crt.2022.1450.
ARID1A Mutation from Targeted Next-Generation Sequencing Predicts Primary Resistance to Gemcitabine and Cisplatin Chemotherapy in Advanced Biliary Tract Cancer
- Affiliations
-
- 1Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, CHA Bundang Medical Center, CHA University School of Medicine, Seongnam, Korea
- 2Division of Medical Oncology, Department of Internal Medicine, Ulsan University Hospital, University of Ulsan College of Medicine, Ulsan, Korea
- 3Department of Internal Medicine, CHA Bundang Medical Center, CHA University School of Medicine, Seongnam, Korea
- 4Division of Medical Oncology, Department of Internal Medicine, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Korea
- 5Department of Pathology, Yonsei University College of Medicine, Seoul, Korea
- 6Department of Radiology, CHA Bundang Medical Center, CHA University School of Medicine, Seongnam, Korea
- KMID: 2547803
- DOI: http://doi.org/10.4143/crt.2022.1450
Abstract
- Purpose
There are clinical unmet needs in predicting therapeutic response and precise strategy for the patient with advanced biliary tract cancer (BTC). We aimed to identify genomic alterations predicting therapeutic response and resistance to gemcitabine and cisplatin (Gem/Cis)-based chemotherapy in advanced BTC.
Materials and Methods
Genomic analysis of advanced BTC multi-institutional cohorts was performed using targeted panel sequencing. Genomic alterations were analyzed integrating patients’ clinicopathologic data, including clinical outcomes of Gem/Cis-based therapy. Significance of genetic alterations was validated using clinical next-generation sequencing (NGS) cohorts from public repositories and drug sensitivity data from cancer cell lines.
Results
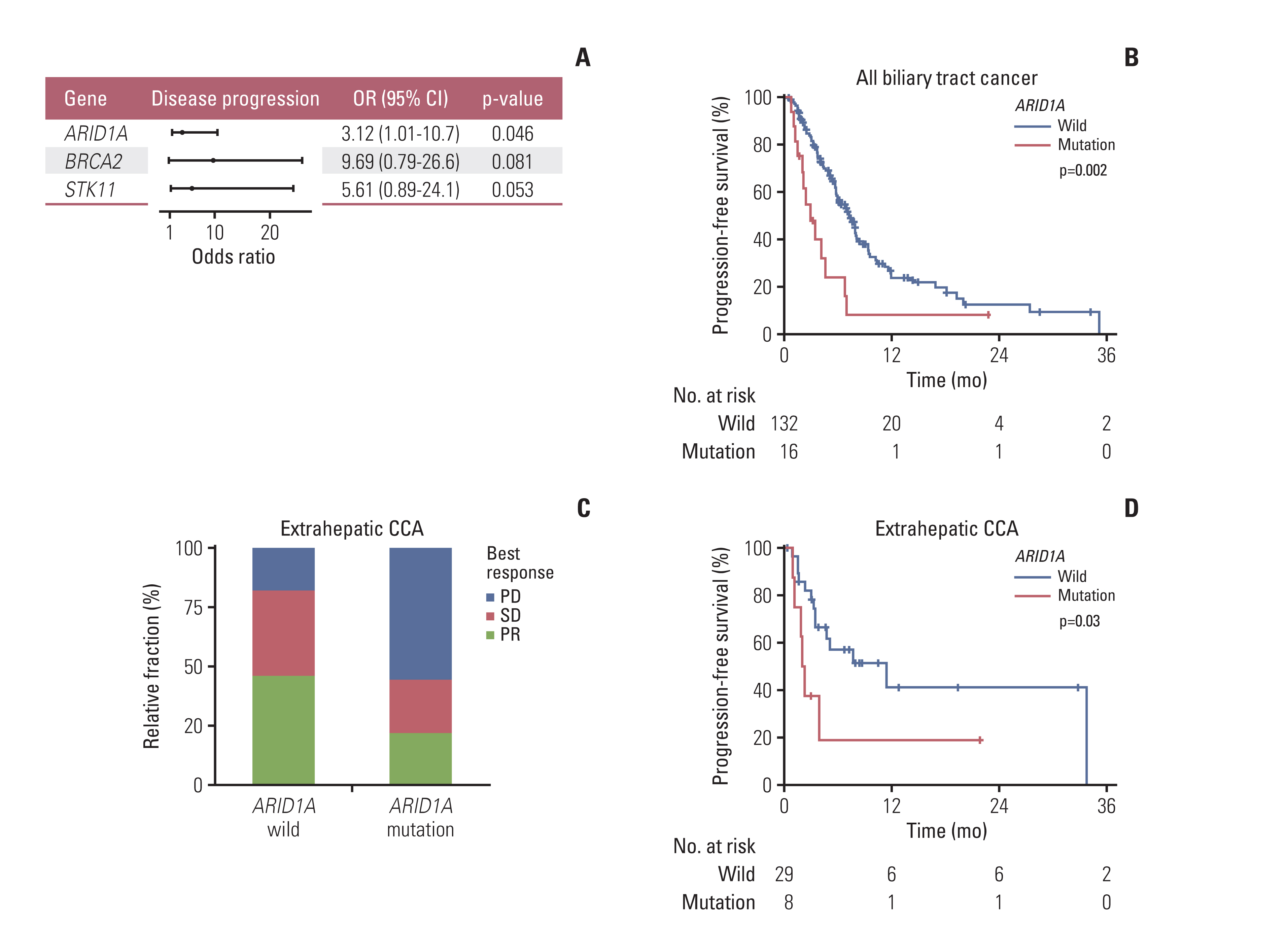
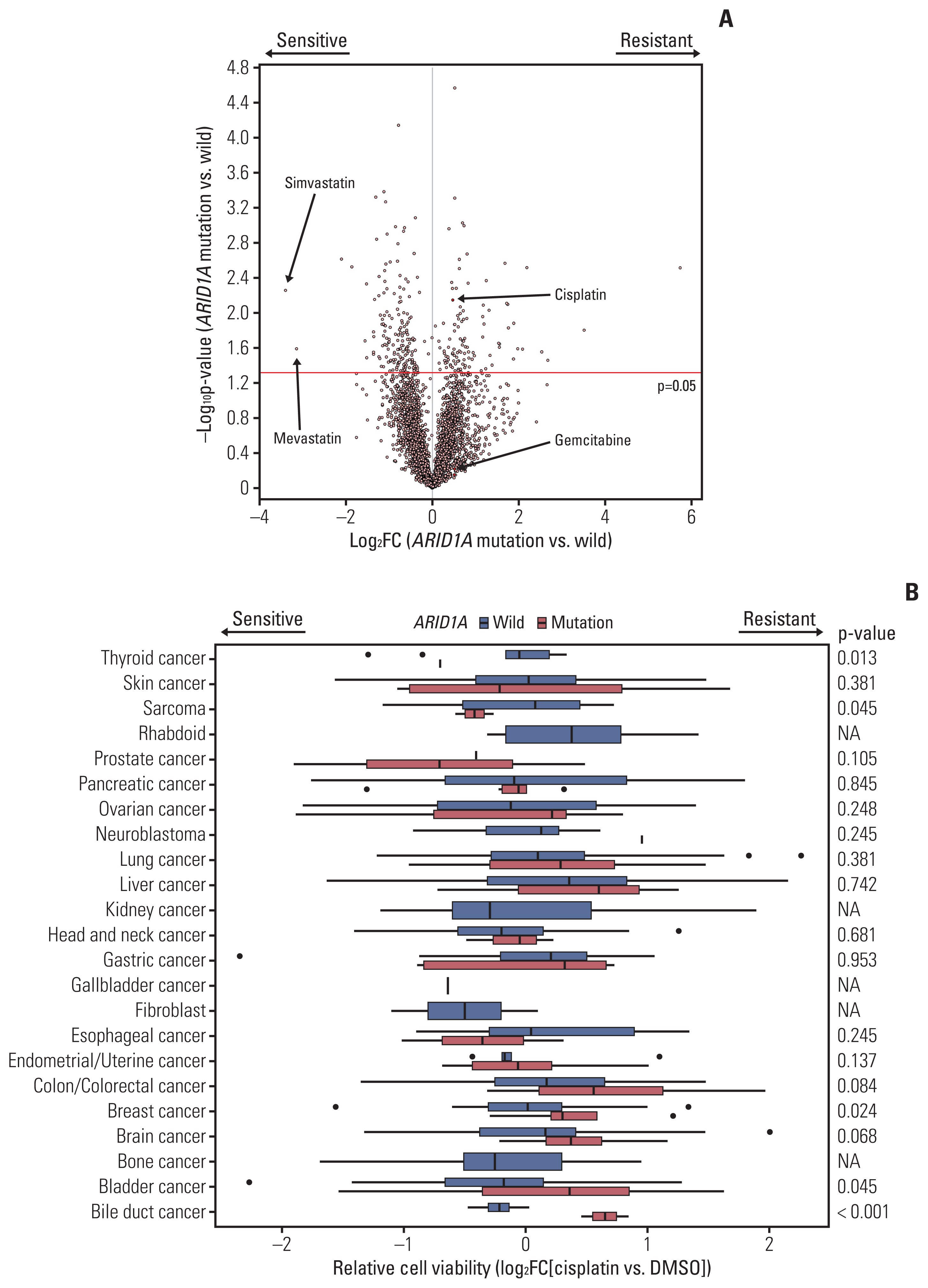
193 BTC patients from three cancer centers were analyzed. Most frequent genomic alterations were TP53 (55.5%), KRAS (22.8%), ARID1A (10.4%) alterations, and ERBB2 amplification (9.8%). Among 177 patients with BTC receiving Gem/Cis-based chemotherapy, ARID1A alteration was the only independent predictive molecular marker of primary resistance showing disease progression for 1st-line chemotherapy in the multivariate regression model (odds ratio, 3.12; p=0.046). In addition, ARID1A alteration was significantly correlated with inferior progression-free survival on Gem/Cis-based chemotherapy in the overall patient population (p=0.033) and in patients with extrahepatic cholangiocarcinoma (CCA) (p=0.041). External validation using public repository NGS revealed that ARID1A mutation was a significant predictor for poor survival in BTC patients. Investigation of multi-OMICs drug sensitivity data from cancer cell lines revealed that cisplatin-resistance was exclusively observed in ARID1A mutant bile duct cancer cells.
Conclusion
Integrative analysis with genomic alterations and clinical outcomes of the first-line Gem/Cis-based chemotherapy in advanced BTC revealed that patients with ARID1Aalterations showed a significant worse clinical outcome, especially in extrahepatic CCA. Well-designed prospective studies are mandatory to validate the predictive role of ARID1Amutation.
Figure
Cited by 1 articles
-
Epidemiology and genomic features of biliary tract cancer and its unique features in Korea
Seonjeong Woo, Youngun Kim, Sohyun Hwang, Hong Jae Chon
J Liver Cancer. 2025;25(1):41-51. doi: 10.17998/jlc.2025.02.27.
Reference
-
References
1. Bridgewater JA, Goodman KA, Kalyan A, Mulcahy MF. Biliary tract cancer: epidemiology, radiotherapy, and molecular profiling. Am Soc Clin Oncol Educ Book. 2016; 35:e194–203.2. Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010; 362:1273–81.
Article3. Valle JW, Lamarca A, Goyal L, Barriuso J, Zhu AX. New horizons for precision medicine in biliary tract cancers. Cancer Discov. 2017; 7:943–62.4. Abou-Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al-Rajabi R, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2020; 21:671–84.
Article5. Zhu AX, Macarulla T, Javle MM, Kelley RK, Lubner SJ, Adeva J, et al. Final overall survival efficacy results of ivosidenib for patients with advanced cholangiocarcinoma with IDH1 mutation: the phase 3 randomized clinical ClarIDHy trial. JAMA Oncol. 2021; 7:1669–77.
Article6. Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020; 31:1491–505.
Article7. Benson AB, D’Angelica MI, Abbott DE, Anaya DA, Anders R, Are C, et al. Hepatobiliary cancers, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021; 19:541–65.8. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017; 19:4–23.
Article9. Lowery MA, Ptashkin R, Jordan E, Berger MF, Zehir A, Capanu M, et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res. 2018; 24:4154–61.
Article10. Chae H, Kim D, Yoo C, Kim KP, Jeong JH, Chang HM, et al. Therapeutic relevance of targeted sequencing in management of patients with advanced biliary tract cancer: DNA damage repair gene mutations as a predictive biomarker. Eur J Cancer. 2019; 120:31–9.
Article11. Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, et al. Genomic spectra of biliary tract cancer. Nat Genet. 2015; 47:1003–10.
Article12. Boscoe AN, Rolland C, Kelley RK. Frequency and prognostic significance of isocitrate dehydrogenase 1 mutations in cholangiocarcinoma: a systematic literature review. J Gastrointest Oncol. 2019; 10:751–65.
Article13. Zhu AX, Borger DR, Kim Y, Cosgrove D, Ejaz A, Alexandrescu S, et al. Genomic profiling of intrahepatic cholangiocarcinoma: refining prognosis and identifying therapeutic targets. Ann Surg Oncol. 2014; 21:3827–34.
Article14. Graham RP, Barr Fritcher EG, Pestova E, Schulz J, Sitailo LA, Vasmatzis G, et al. Fibroblast growth factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Hum Pathol. 2014; 45:1630–8.
Article15. Ross JS, Wang K, Gay L, Al-Rohil R, Rand JV, Jones DM, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist. 2014; 19:235–42.
Article16. Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep. 2017; 19:2878–80.17. Bruno R, Fontanini G. Next generation sequencing for gene fusion analysis in lung cancer: a literature review. Diagnostics (Basel). 2020; 10:521.
Article18. Yoon JG, Kim MH, Jang M, Kim H, Hwang HK, Kang CM, et al. Molecular characterization of biliary tract cancer predicts chemotherapy and programmed death 1/programmed death-ligand 1 blockade responses. Hepatology. 2021; 74:1914–31.
Article19. Montal R, Sia D, Montironi C, Leow WQ, Esteban-Fabro R, Pinyol R, et al. Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J Hepatol. 2020; 73:315–27.
Article20. Pavlidou EN, Balis V. Diagnostic significance and prognostic role of the ARID1A gene in cancer outcomes (review). World Acad Sci J. 2020; 2:49–64.
Article21. Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011; 43:1219–23.
Article22. Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010; 363:1532–43.
Article23. Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A. 2012; 109:E252–9.
Article24. Luchini C, Veronese N, Solmi M, Cho H, Kim JH, Chou A, et al. Prognostic role and implications of mutation status of tumor suppressor gene ARID1A in cancer: a systematic review and meta-analysis. Oncotarget. 2015; 6:39088–97.
Article25. Berns K, Sonnenblick A, Gennissen A, Brohee S, Hijmans EM, Evers B, et al. Loss of ARID1A activates ANXA1, which serves as a predictive biomarker for trastuzumab resistance. Clin Cancer Res. 2016; 22:5238–48.26. Javle M, Bekaii-Saab T, Jain A, Wang Y, Kelley RK, Wang K, et al. Biliary cancer: Utility of next-generation sequencing for clinical management. Cancer. 2016; 122:3838–47.
Article27. Conci S, Ruzzenente A, Simbolo M, Bagante F, Rusev B, Isa G, et al. Multigene mutational profiling of biliary tract cancer is related to the pattern of recurrence in surgically resected patients. Updates Surg. 2020; 72:119–28.
Article28. Xu SF, Guo Y, Zhang X, Zhu XD, Fan N, Zhang ZL, et al. Somatic mutation profiling of intrahepatic cholangiocarcinoma: comparison between primary and metastasis tumor tissues. J Oncol. 2020; 2020:5675020.
Article29. McGregor GH, Campbell AD, Fey SK, Tumanov S, Sumpton D, Blanco GR, et al. Targeting the metabolic response to statin-mediated oxidative stress produces a synergistic antitumor response. Cancer Res. 2020; 80:175–88.
Article30. Wu S, Fukumoto T, Lin J, Nacarelli T, Wang Y, Ong D, et al. Targeting glutamine dependence through GLS1 inhibition suppresses ARID1A-inactivated clear cell ovarian carcinoma. Nat Cancer. 2021; 2:189–200.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Current status of chemotherapy for the treatment of advanced biliary tract cancer
- Chemotherapy for Biliary Tract Cancer
- Novel Palliative Chemotherapy for Cholangiocarcinoma
- Recent Advances in Immunotherapy and Targeted Therapy for Biliary Tract Cancer
- Real-World Outcomes of Gemcitabine, Cisplatin, and Nab-Paclitaxel Chemotherapy Regimen for Advanced Biliary Tract Cancer: A Propensity Score-Matched Analysis