Int J Stem Cells.
2023 Aug;16(3):356-362. 10.15283/ijsc23047.
Measuring Glutathione Regeneration Capacity in Stem Cells
- Affiliations
-
- 1Department of Pharmacy, College of Pharmacy, Jeju Research Institute of Pharmaceutical Sciences, Jeju National University, Jeju, Korea
- 2Interdisciplinary Graduate Program in Advanced Convergence Technology and Science, Bio-Health Materials Core-Facility Center and Practical Translational Research Center, Jeju National University, Jeju, Korea
- KMID: 2545234
- DOI: http://doi.org/10.15283/ijsc23047
Abstract
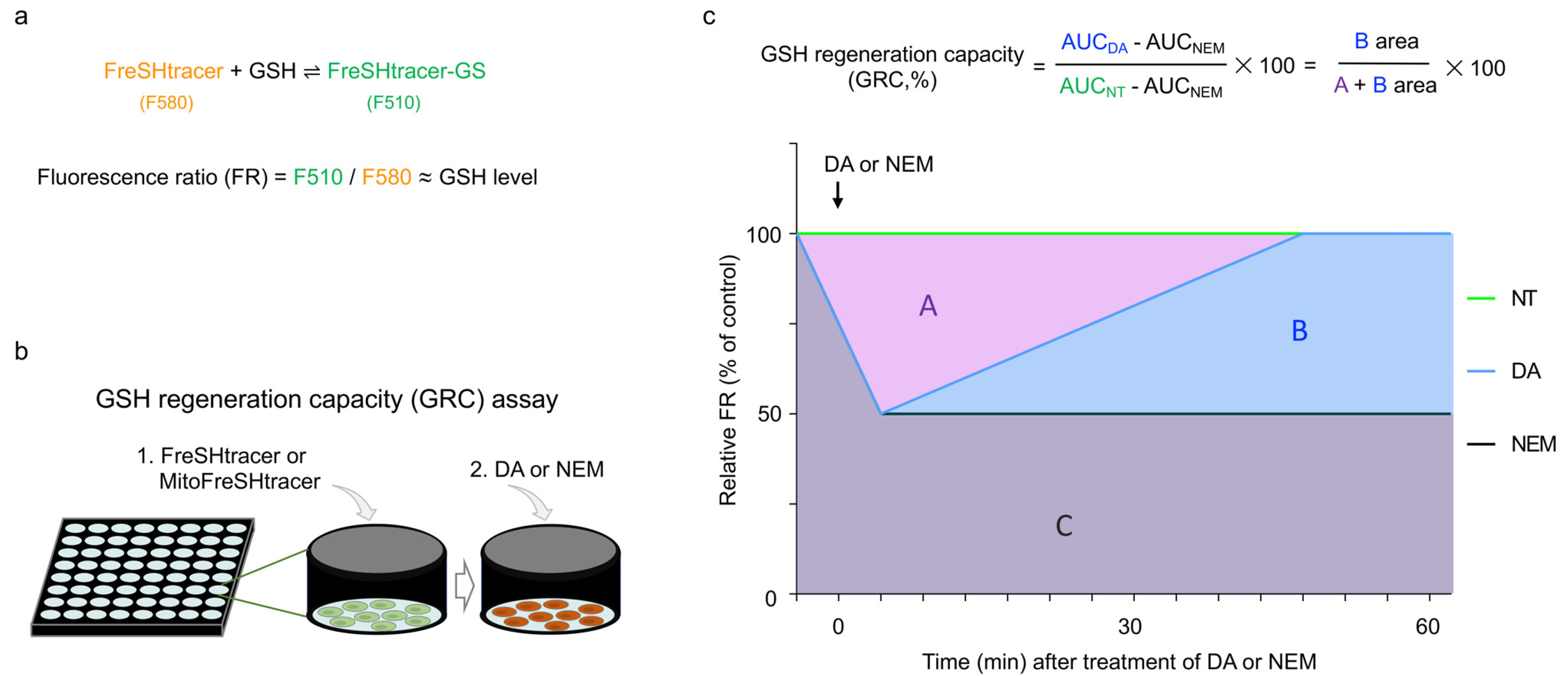
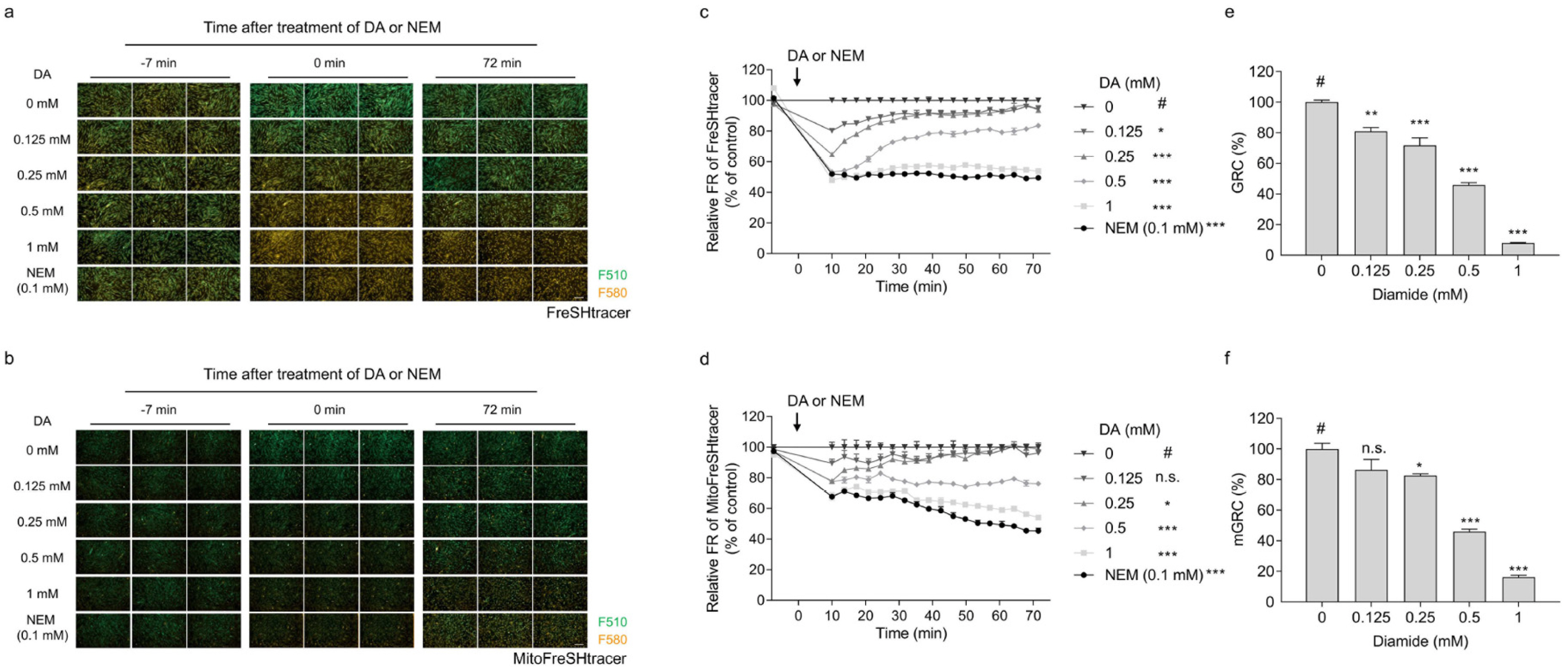
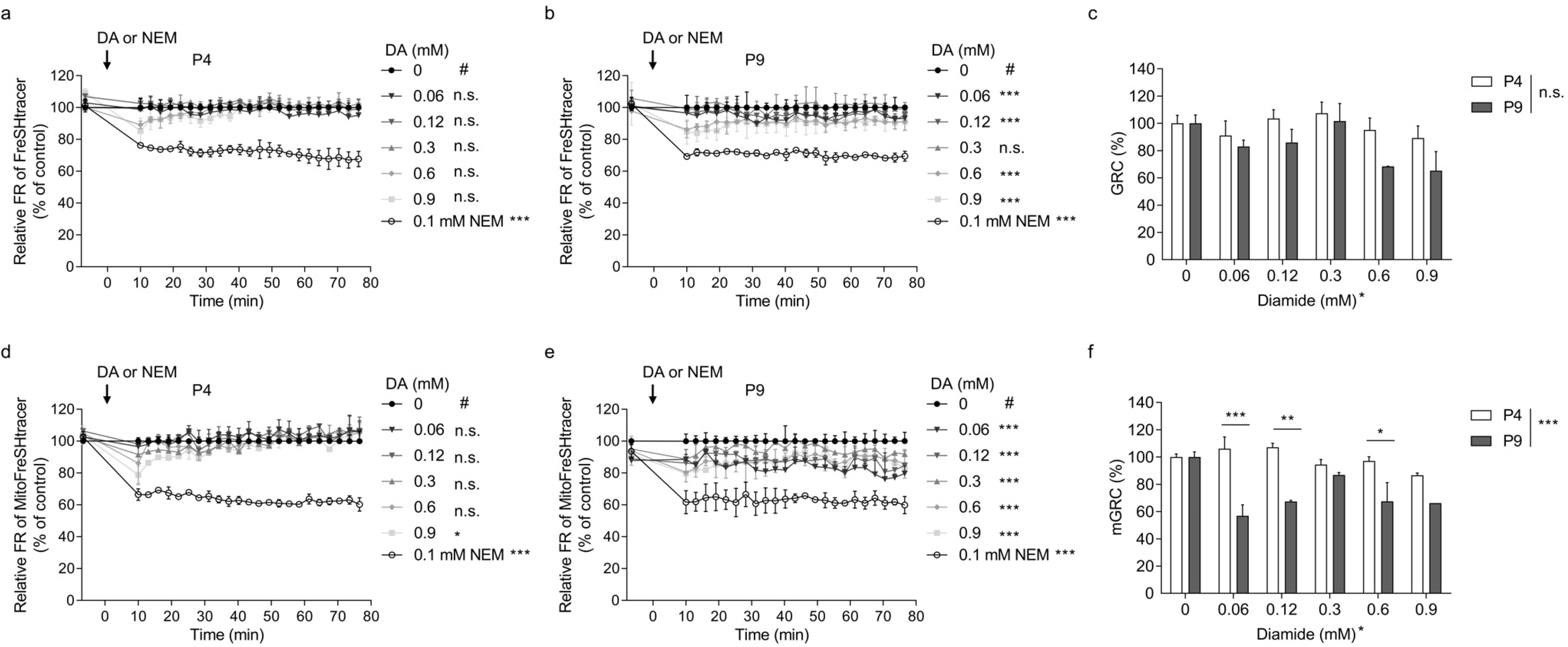
- Glutathione (GSH) is a chief cellular antioxidant, affecting stem cell functions. The cellular GSH level is dynamically altered by the redox buffering system and transcription factors, including NRF2. Additionally, GSH is differentially regulated in each organelle. We previously reported a protocol for monitoring the real-time GSH levels in live stem cells using the reversible GSH sensor FreSHtracer. However, GSH-based stem cell analysis needs be comprehensive and organelle-specific. Hence, in this study, we demonstrate a detailed protocol to measure the GSH regeneration capacity (GRC) in living stem cells by measuring the intensities of the FreSHtracer and the mitochondrial GSH sensor MitoFreSHtracer using a high-content screening confocal microscope. This protocol typically analyses the GRC in approximately 4 h following the seeding of the cells onto plates. This protocol is simple and quantitative. With some minor modifications, it can be employed flexibly to measure the GRC for the whole-cell area or just the mitochondria in all adherent mammalian stem cells.
Figure
Reference
-
References
1. Lu SC. 2013; Glutathione synthesis. Biochim Biophys Acta. 1830:3143–3153. DOI: 10.1016/j.bbagen.2012.09.008. PMID: 22995213. PMCID: PMC3549305.
Article2. Winterbourn CC, Hampton MB. 2008; Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 45:549–561. DOI: 10.1016/j.freeradbiomed.2008.05.004. PMID: 18544350.
Article3. Jeong EM, Yoon JH, Lim J, Shin JW, Cho AY, Heo J, Lee KB, Lee JH, Lee WJ, Kim HJ, Son YH, Lee SJ, Cho SY, Shin DM, Choi K, Kim IG. 2018; Real-time monitoring of glutathione in living cells reveals that high glutathione levels are required to maintain stem cell function. Stem Cell Reports. 10:600–614. DOI: 10.1016/j.stemcr.2017.12.007. PMID: 29307581. PMCID: PMC5830891.
Article4. Lim J, Heo J, Ju H, Shin JW, Kim Y, Lee S, Yu HY, Ryu CM, Yun H, Song S, Hong KS, Chung HM, Kim HR, Roe JS, Choi K, Kim IG, Jeong EM, Shin DM. 2020; Glutathione dynamics determine the therapeutic efficacy of mesenchymal stem cells for graft-versus-host disease via CREB1-NRF2 pathway. Sci Adv. 6:eaba1334. DOI: 10.1126/sciadv.aba1334. PMID: 32490200. PMCID: PMC7239701.
Article5. Lim J, Heo J, Yu HY, Yun H, Lee S, Ju H, Nam YJ, Jeong SM, Lee J, Cho YS, Choo MS, Jeong EM, Ryu CM, Shin DM. 2021; Small-sized mesenchymal stem cells with high glutathione dynamics show improved therapeutic potency in graft-versus-host disease. Clin Transl Med. 11:e476. DOI: 10.1002/ctm2.476.
Article6. Ribas V, García-Ruiz C, Fernández-Checa JC. 2014; Glutathione and mitochondria. Front Pharmacol. 5:151. DOI: 10.3389/fphar.2014.00151. PMID: 25024695. PMCID: PMC4079069.
Article7. Marí M, Morales A, Colell A, García-Ruiz C, Kaplowitz N, Fernández-Checa JC. 2013; Mitochondrial glutathione: features, regulation and role in disease. Biochim Biophys Acta. 1830:3317–3328. DOI: 10.1016/j.bbagen.2012.10.018. PMID: 23123815. PMCID: PMC3578987.
Article8. Cho AY, Choi K. 2012; A coumarin-based fluorescence sensor for the reversible detection of thiols. Chem Lett. 41:1611–1612. DOI: 10.1246/cl.2012.1611.
Article9. Jeong EM, Shin JW, Lim J, Kim JH, Kang H, Yin Y, Kim HM, Kim Y, Kim SG, Kang HS, Shin DM, Choi K, Kim IG. 2019; Monitoring glutathione dynamics and heterogeneity in living stem cells. Int J Stem Cells. 12:367–379. DOI: 10.15283/ijsc18151. PMID: 30836726. PMCID: PMC6657947.
Article10. Porcelli AM, Ghelli A, Zanna C, Pinton P, Rizzuto R, Rugolo M. 2005; pH difference across the outer mitochondrial membrane measured with a green fluorescent protein mutant. Biochem Biophys Res Commun. 326:799–804. DOI: 10.1016/j.bbrc.2004.11.105. PMID: 15607740.
Article