Korean J Physiol Pharmacol.
2023 Jul;27(4):333-344. 10.4196/kjpp.2023.27.4.333.
Circ-SNX27 sponging miR-375/RPN1 axis contributes to hepatocellular carcinoma progression
- Affiliations
-
- 1Surgical Department, Wuhan Hospital of Traditional Chinese Medicine, Wuhan 430000, China
- 2Department of Spleen Stomach Disease and Hepatobiliary Disease, Wuhan Hospital of Traditional Chinese Medicine, Wuhan 430000, China
- KMID: 2544135
- DOI: http://doi.org/10.4196/kjpp.2023.27.4.333
Abstract
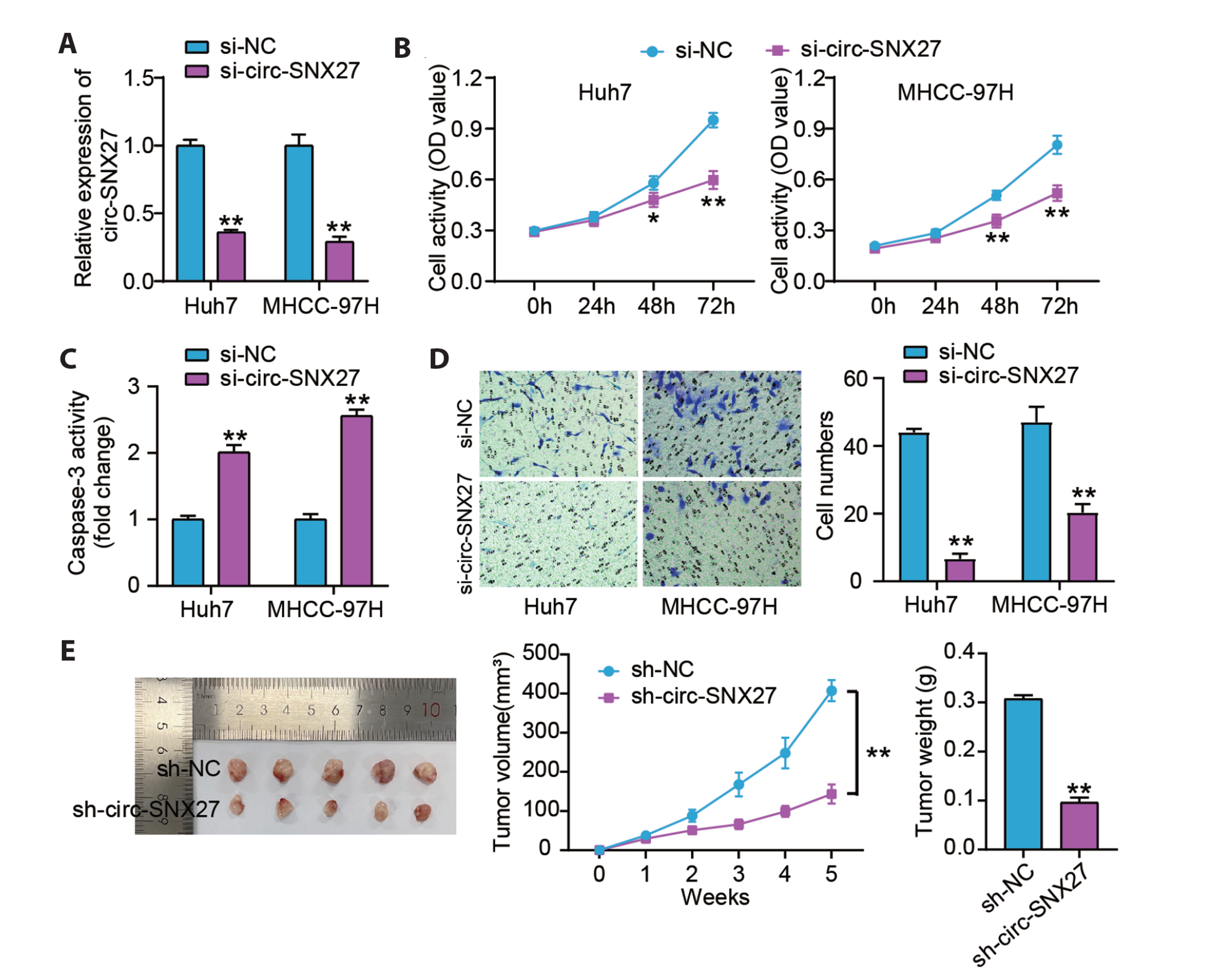
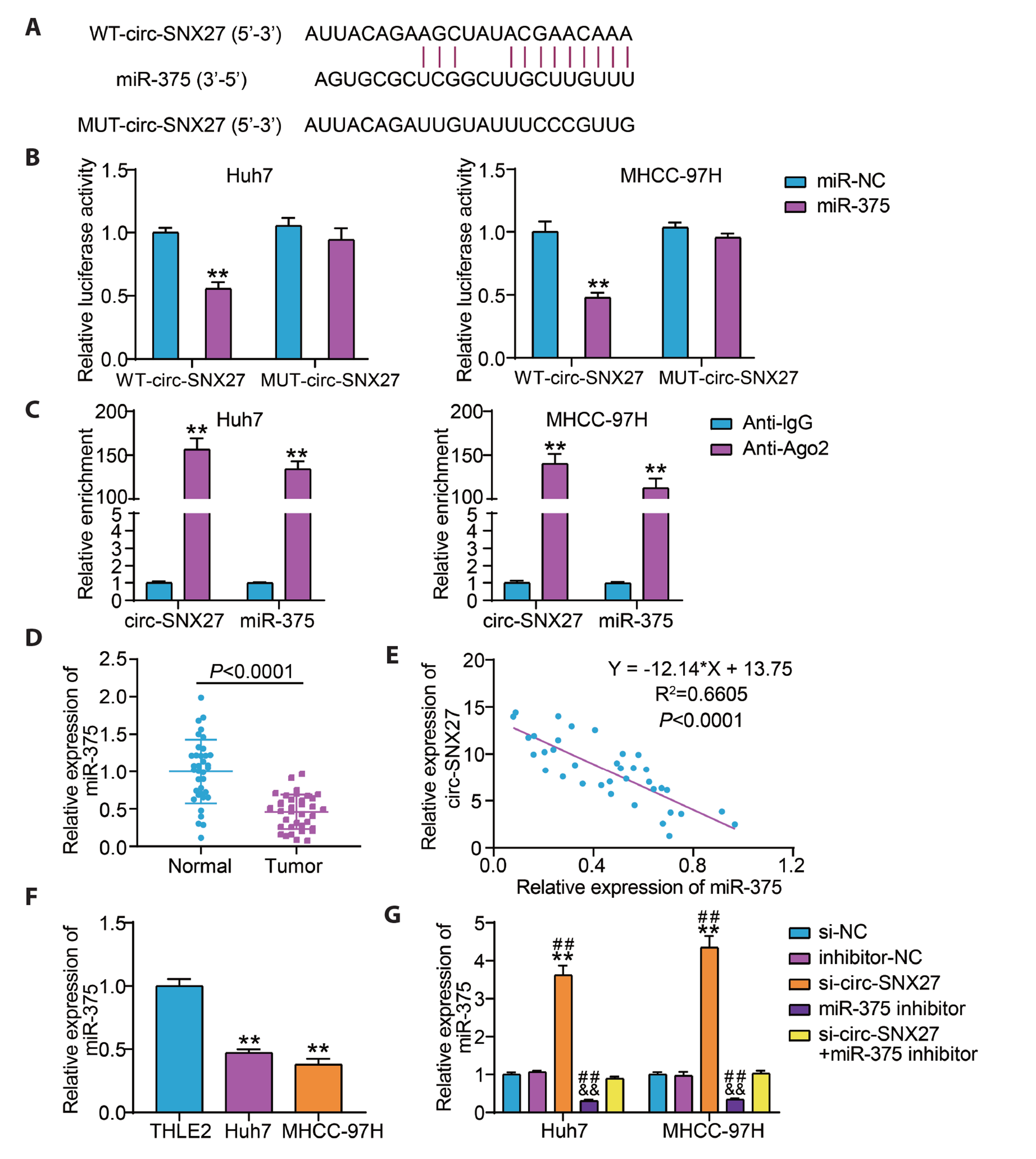
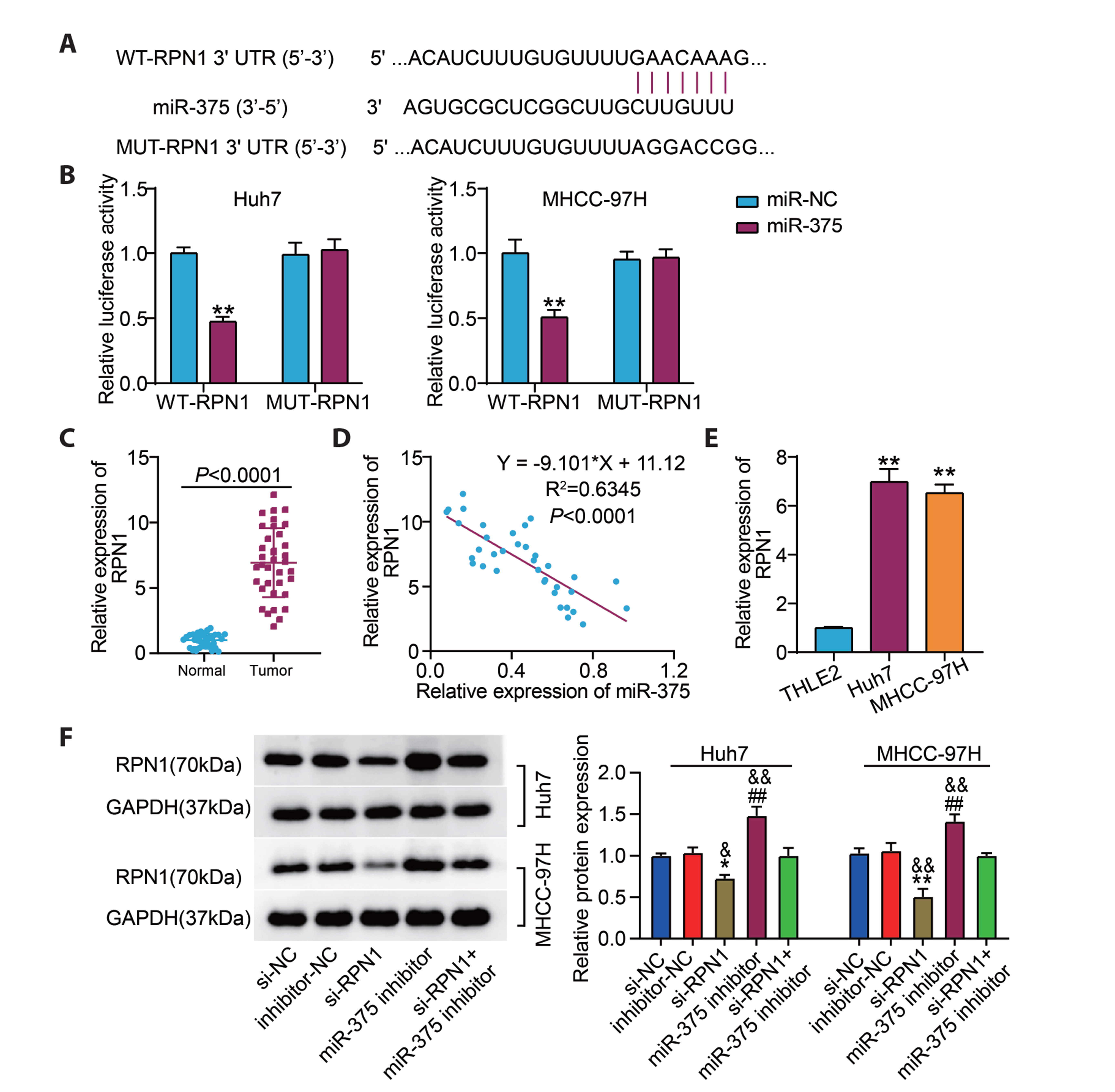
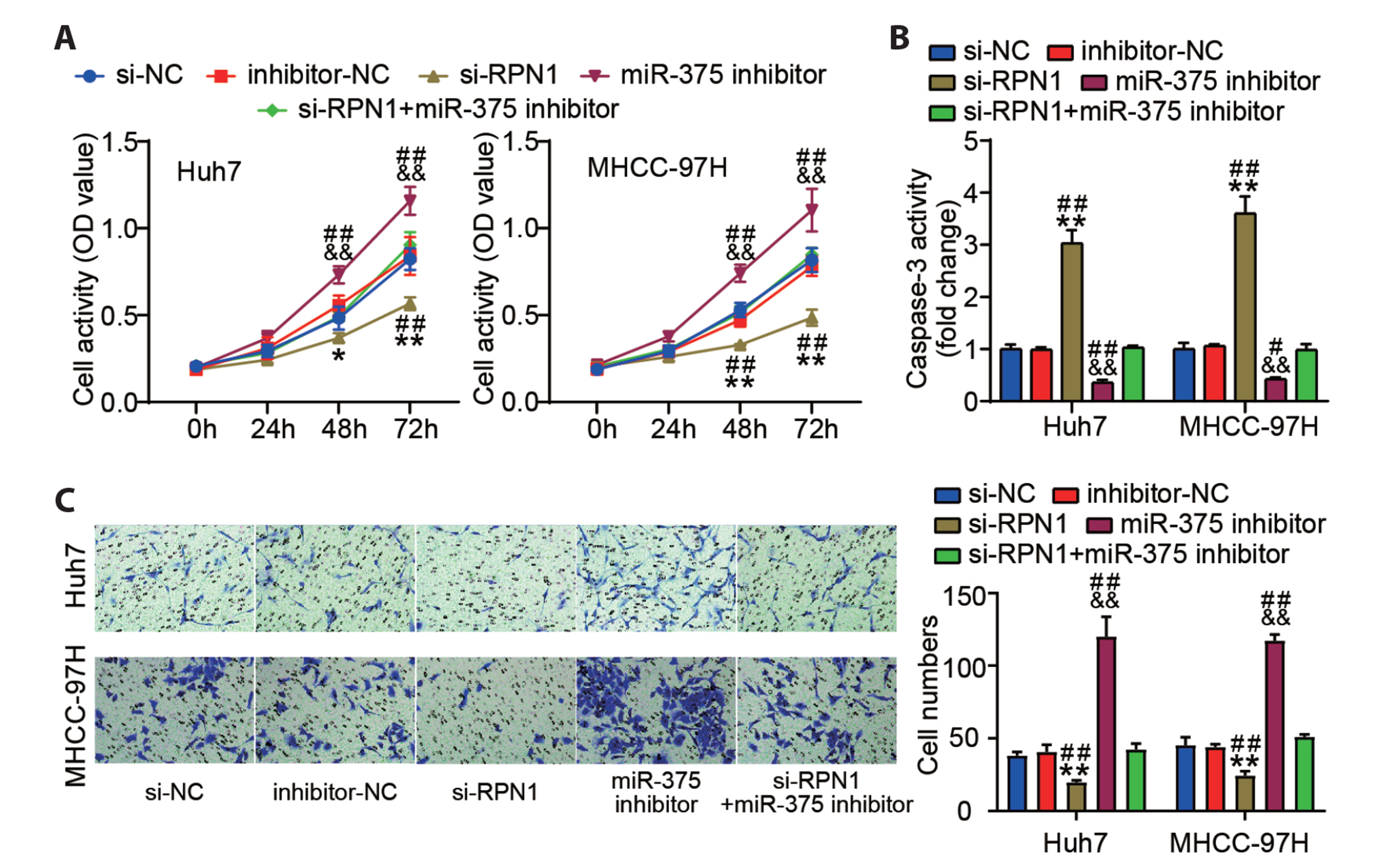
- Hepatocellular carcinoma (HCC) is a prevalent malignant tumor with high fatality. It has yet to be reported whether circ-SNX27 can affect the progression of HCC. This study attempted to analyze circ-SNX27’s precise role and underlying mechanisms in HCC. HCC cell lines and tumor specimens from HCC patients were analyzed using quantitative real-time PCR and Western blotting to quantify the expressions of circ-SNX27, miR-375, and ribophorin I (RPN1). Cell invasion and cell counting kit 8 experiments were conducted for the evaluation of HCC cell invasion and proliferation. Caspase-3 Activity Assay Kit was utilized to gauge the caspase-3 activity. Luciferase reporter and RNA immunoprecipitation assays were executed to ascertain the relationships among miR-375, circ-SNX27, and RPN1. To determine how circ-SNX27 knockdown affects the growth of HCC xenografts in vivo, tumor-bearing mouse models were constructed. Elevated expressions of circ-SNX27 and RPN1 as well as a reduced miR-375 expression were observed among HCC cells and HCC patient tumor specimens. Knocking-down circ-SNX27 in HCC cells abated their proliferative and invasive abilities but raised their caspase-3 activity. Moreover, the poor levels of circ-SNX27 inhibited HCC tumor growth among the mice. Circ-SNX27 enhanced RPN1 by competitively binding with miR-375. Silencing miR-375 in HCC cells promoted their malignant phenotypes. Nonetheless, the promotive effect of miR-375 silencing was reversible via the knockdown of circ-SNX27 or RPN1. This research demonstrated that circ-SNX27 accelerated the progression of HCC by modulating the miR-375/RPN1 axis. This is indicative of circ-SNX27’s potential as a target for the treatment of HCC.
Figure
Reference
-
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. 2021; Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. DOI: 10.3322/caac.21660. PMID: 33538338.
Article2. Eugen K. 2020; Current treatment options for hepatocellular carcinoma. Klin Onkol. 33(Supplementum 3):20–25. DOI: 10.14735/amko20203S20. PMID: 33213161.
Article3. Hartke J, Johnson M, Ghabril M. 2017; The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 34:153–159. DOI: 10.1053/j.semdp.2016.12.011. PMID: 28108047.
Article4. Pascual S, Herrera I, Irurzun J. 2016; New advances in hepatocellular carcinoma. World J Hepatol. 8:421–438. DOI: 10.4254/wjh.v8.i9.421. PMID: 27028578. PMCID: PMC4807304.
Article5. Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. 2020; Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 19:172. DOI: 10.1186/s12943-020-01286-3. PMID: 33317550. PMCID: PMC7734744. PMID: d632b21718e94b679d98a2fde221e564.
Article6. Qiu L, Xu H, Ji M, Shang D, Lu Z, Wu Y, Tu Z, Liu H. 2019; Circular RNAs in hepatocellular carcinoma: biomarkers, functions and mechanisms. Life Sci. 231:116660. DOI: 10.1016/j.lfs.2019.116660. PMID: 31319086.
Article7. Qiu L, Wang T, Ge Q, Xu H, Wu Y, Tang Q, Chen K. 2019; Circular RNA signature in hepatocellular carcinoma. J Cancer. 10:3361–3372. DOI: 10.7150/jca.31243. PMID: 31293639. PMCID: PMC6603403.
Article8. Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, Mao L, Luo CB, Yu SY, Huang XW, Cao Y, Fan J, Zhou J. 2020; Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. 72:906–922. DOI: 10.1002/hep.31068. PMID: 31838741.
Article9. Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, Cai JB, Yang X, Fan J, Ke AW, Zhou J, Shi GM. 2020; Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer. 19:92. DOI: 10.1186/s12943-020-01213-6. PMID: 32430013. PMCID: PMC7236145. PMID: c04084b82de94621b6820d97eb69f9a9.
Article10. Zhang PF, Wei CY, Huang XY, Peng R, Yang X, Lu JC, Zhang C, Gao C, Cai JB, Gao PT, Gao DM, Shi GM, Ke AW, Fan J. 2019; Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. 18:105. DOI: 10.1186/s12943-019-1031-1. PMID: 31153371. PMCID: PMC6545035. PMID: 0941232519964eb28e6de3caea9e3619.
Article11. Oura K, Morishita A, Masaki T. 2020; Molecular and functional roles of MicroRNAs in the progression of hepatocellular carcinoma- a review. Int J Mol Sci. 21:8362. DOI: 10.3390/ijms21218362. PMID: 33171811. PMCID: PMC7664704. PMID: 6b8ff7c912b040bbb0dcb0b70fc64189.
Article12. Lin H, Zhang R, Wu W, Lei L. 2021; miR-4454 promotes hepatic carcinoma progression by targeting Vps4A and Rab27A. Oxid Med Cell Longev. 2021:9230435. DOI: 10.1155/2021/9230435. PMID: 34777698. PMCID: PMC8580624.
Article13. Hirao A, Sato Y, Tanaka H, Nishida K, Tomonari T, Hirata M, Bando M, Kida Y, Tanaka T, Kawaguchi T, Wada H, Taniguchi T, Okamoto K, Miyamoto H, Muguruma N, Tanahashi T, Takayama T. 2021; MiR-125b-5p is involved in sorafenib resistance through ataxin-1-mediated epithelial-mesenchymal transition in hepatocellular carcinoma. Cancers (Basel). 13:4917. DOI: 10.3390/cancers13194917. PMID: 34638401. PMCID: PMC8508441. PMID: de7840a3314e4a9ab1c3b4ebeda2b913.
Article14. Li D, Wang T, Sun FF, Feng JQ, Peng JJ, Li H, Wang C, Wang D, Liu Y, Bai YD, Shi ML, Zhang T. 2021; MicroRNA-375 represses tumor angiogenesis and reverses resistance to sorafenib in hepatocarcinoma. Cancer Gene Ther. 28:126–140. DOI: 10.1038/s41417-020-0191-x. PMID: 32616906. PMCID: PMC7886652.
Article15. Wang C, Luo J, Chen Z, Ye M, Hong Y, Liu J, Nie J, Zhao Q, Chang Y. 2021; MiR-375 impairs the invasive capabilities of hepatoma cells by targeting HIF1α under hypoxia. Dig Dis Sci. 66:493–502. DOI: 10.1007/s10620-020-06202-9. PMID: 32215815.
Article16. Takeda K, Qin SY, Matsumoto N, Yamamoto K. 2014; Association of malectin with ribophorin I is crucial for attenuation of misfolded glycoprotein secretion. Biochem Biophys Res Commun. 454:436–440. DOI: 10.1016/j.bbrc.2014.10.102. PMID: 25451265.
Article17. Ding J, Xu J, Deng Q, Ma W, Zhang R, He X, Liu S, Zhang L. 2021; Knockdown of Oligosaccharyltransferase subunit ribophorin 1 induces endoplasmic-reticulum-stress-dependent cell apoptosis in breast cancer. Front Oncol. 11:722624. DOI: 10.3389/fonc.2021.722624. PMID: 34778038. PMCID: PMC8578895. PMID: 6da03cac95164c21ab93aa445f99c353.
Article18. Liu D, Li L, Wang L, Wang C, Hu X, Jiang Q, Wang X, Xue G, Liu Y, Xue D. 2021; Recognition of DNA methylation molecular features for diagnosis and prognosis in gastric cancer. Front Genet. 12:758926. DOI: 10.3389/fgene.2021.758926. PMID: 34745226. PMCID: PMC8566671. PMID: 9fa9baea30a043f688655922bef75ce1.
Article19. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. 2016; CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 13:34–42. DOI: 10.1080/15476286.2015.1128065. PMID: 26669964. PMCID: PMC4829301.
Article20. Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, Shu Y. 2019; CircRNAs in cancer metabolism: a review. J Hematol Oncol. 12:90. DOI: 10.1186/s13045-019-0776-8. PMID: 31484561. PMCID: PMC6727394. PMID: a8a94aef9e8d402fbf07ab6c1955359b.
Article21. Xiong DD, Dang YW, Lin P, Wen DY, He RQ, Luo DZ, Feng ZB, Chen G. 2018; A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 16:220. DOI: 10.1186/s12967-018-1593-5. PMID: 30092792. PMCID: PMC6085698. PMID: dc69d800dd6349869c0b534291b49bb1.
Article22. Ji Y, Yang S, Yan X, Zhu L, Yang W, Yang X, Yu F, Shi L, Zhu X, Lu Y, Zhang C, Lu H, Zhang F. 2021; CircCRIM1 promotes hepatocellular carcinoma proliferation and angiogenesis by sponging miR-378a-3p and regulating SKP2 expression. Front Cell Dev Biol. 9:796686. DOI: 10.3389/fcell.2021.796686. PMID: 34869393. PMCID: PMC8634842. PMID: fecaed6fef494100bbadfdad8baca916.
Article23. Huang C, Yu W, Wang Q, Huang T, Ding Y. 2021; CircANTXR1 contributes to the malignant progression of hepatocellular carcinoma by promoting proliferation and metastasis. J Hepatocell Carcinoma. 8:1339–1353. DOI: 10.2147/JHC.S317256. PMID: 34786378. PMCID: PMC8590609.
Article24. Xing W, Zhou PC, Zhang HY, Chen LM, Zhou YM, Cui XF, Liu ZG. 2022; Circular RNA circ_GLIS2 suppresses hepatocellular carcinoma growth and metastasis. Liver Int. 42:682–695. DOI: 10.1111/liv.15097. PMID: 34743403.
Article25. Zhang L, Zhang J, Li P, Li T, Zhou Z, Wu H. 2022; Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death Dis. 13:32. DOI: 10.1038/s41419-021-04345-9. PMID: 35013102. PMCID: PMC8748962. PMID: 502fa7fb80b04c35ac3ca8aec42a0800.
Article26. Li D, Zhang J, Yang J, Wang J, Zhang R, Li J, Zhang R. 2021; CircMTO1 suppresses hepatocellular carcinoma progression via the miR-541-5p/ZIC1 axis by regulating Wnt/β-catenin signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis. 13:12. DOI: 10.1038/s41419-021-04464-3. PMID: 34930906. PMCID: PMC8688446. PMID: d4fa7a43ad5d47aa89dbbfd957771eab.
Article27. Chang Y, Yan W, He X, Zhang L, Li C, Huang H, Nace G, Geller DA, Lin J, Tsung A. 2012; miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 143:177–187.e8. DOI: 10.1053/j.gastro.2012.04.009. PMID: 22504094.
Article28. Li L, Xiao C, He K, Xiang G. 2021; Circ_0072088 promotes progression of hepatocellular carcinoma by activating JAK2/STAT3 signaling pathway via miR-375. IUBMB Life. 73:1153–1165. DOI: 10.1002/iub.2520. PMID: 34148288.
Article29. Fan YP, Liao JZ, Lu YQ, Tian DA, Ye F, Zhao PX, Xiang GY, Tang WX, He XX. 2017; MiR-375 and doxorubicin co-delivered by liposomes for combination therapy of hepatocellular carcinoma. Mol Ther Nucleic Acids. 7:181–189. DOI: 10.1016/j.omtn.2017.03.010. PMID: 28624193. PMCID: PMC5415965. PMID: a5e56e9582a44791a292feb19a4e6314.
Article30. Zhao P, Wu S, Cheng Y, You J, Chen Y, Li M, He C, Zhang X, Yang T, Lu Y, Lee RJ, He X, Xiang G. 2017; MiR-375 delivered by lipid-coated doxorubicin-calcium carbonate nanoparticles overcomes chemoresistance in hepatocellular carcinoma. Nanomedicine. 13:2507–2516. DOI: 10.1016/j.nano.2017.05.010. PMID: 28577837.
Article31. Milde-Langosch K, Karn T, Schmidt M, zu Eulenburg C, Oliveira-Ferrer L, Wirtz RM, Schumacher U, Witzel I, Schütze D, Müller V. 2014; Prognostic relevance of glycosylation-associated genes in breast cancer. Breast Cancer Res Treat. 145:295–305. DOI: 10.1007/s10549-014-2949-z. PMID: 24737166.
Article32. Haltrich I, Kost-Alimova M, Kovács G, Klein G, Fekete G, Imreh S. 2006; Multipoint interphase FISH analysis of chromosome 3 abnormalities in 28 childhood AML patients. Eur J Haematol. 76:124–133. DOI: 10.1111/j.1600-0609.2005.00576.x. PMID: 16405433.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Circular RNA-ZFR Inhibited Cell Proliferation and Promoted Apoptosis in Gastric Cancer by Sponging miR-130a/miR-107 and Modulating PTEN
- XIST Induced by JPX Suppresses Hepatocellular Carcinoma by Sponging miR-155-5p
- RETRACTION: Circular RNA-ZFR Inhibited Cell Proliferation and Promoted Apoptosis in Gastric Cancer by Sponging miR-130a/miR-107 and Modulating PTEN
- MicroRNA Expression in Plasma of Esophageal Squamous Cell Carcinoma Patients
- MicroRNA-370 Regulates Cellepithelial-Mesenchymal Transition, Migration, Invasion, and Prognosis of Hepatocellular Carcinoma by Targeting GUCD1