Healthc Inform Res.
2023 Apr;29(2):132-144. 10.4258/hir.2023.29.2.132.
Standardized Database of 12-Lead Electrocardiograms with a Common Standard for the Promotion of Cardiovascular Research: KURIAS-ECG
- Affiliations
-
- 1Korea University Research Institute for Medical Bigdata Science, Korea University College of Medicine, Seoul, Korea
- 2Department of Biostatistics, Korea University College of Medicine, Seoul, Korea
- 3School of Computer Science and Information Engineering, The Catholic University of Korea, Bucheon, Korea
- 4Department of Cardiology, Cardiovascular Center, Korea University College of Medicine, Seoul, Korea
- 5Korea University Research Institute for Healthcare Service Innovation, Korea University College of Medicine, Seoul, Korea
- 6Department of Emergency Medicine, Korea University Anam Hospital, Korea University College of Medicine, Seoul, Korea
- 7Department of Medical Informatics, Korea University College of Medicine, Seoul, Korea
- KMID: 2542139
- DOI: http://doi.org/10.4258/hir.2023.29.2.132
Abstract
Objectives
Electrocardiography (ECG)-based diagnosis by experts cannot maintain uniform quality because individual differences may occur. Previous public databases can be used for clinical studies, but there is no common standard that would allow databases to be combined. For this reason, it is difficult to conduct research that derives results by combining databases. Recent commercial ECG machines offer diagnoses similar to those of a physician. Therefore, the purpose of this study was to construct a standardized ECG database using computerized diagnoses.
Methods
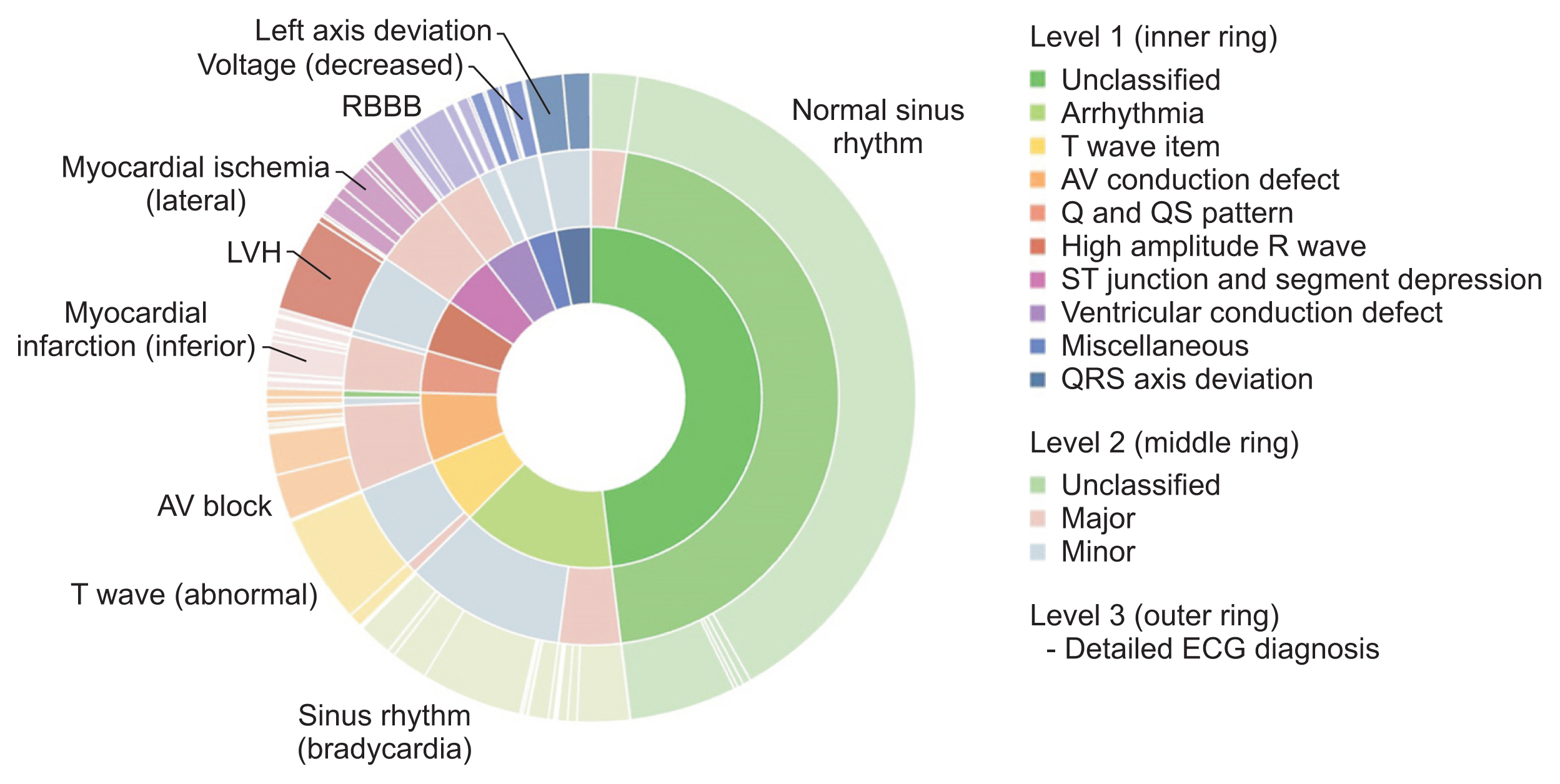
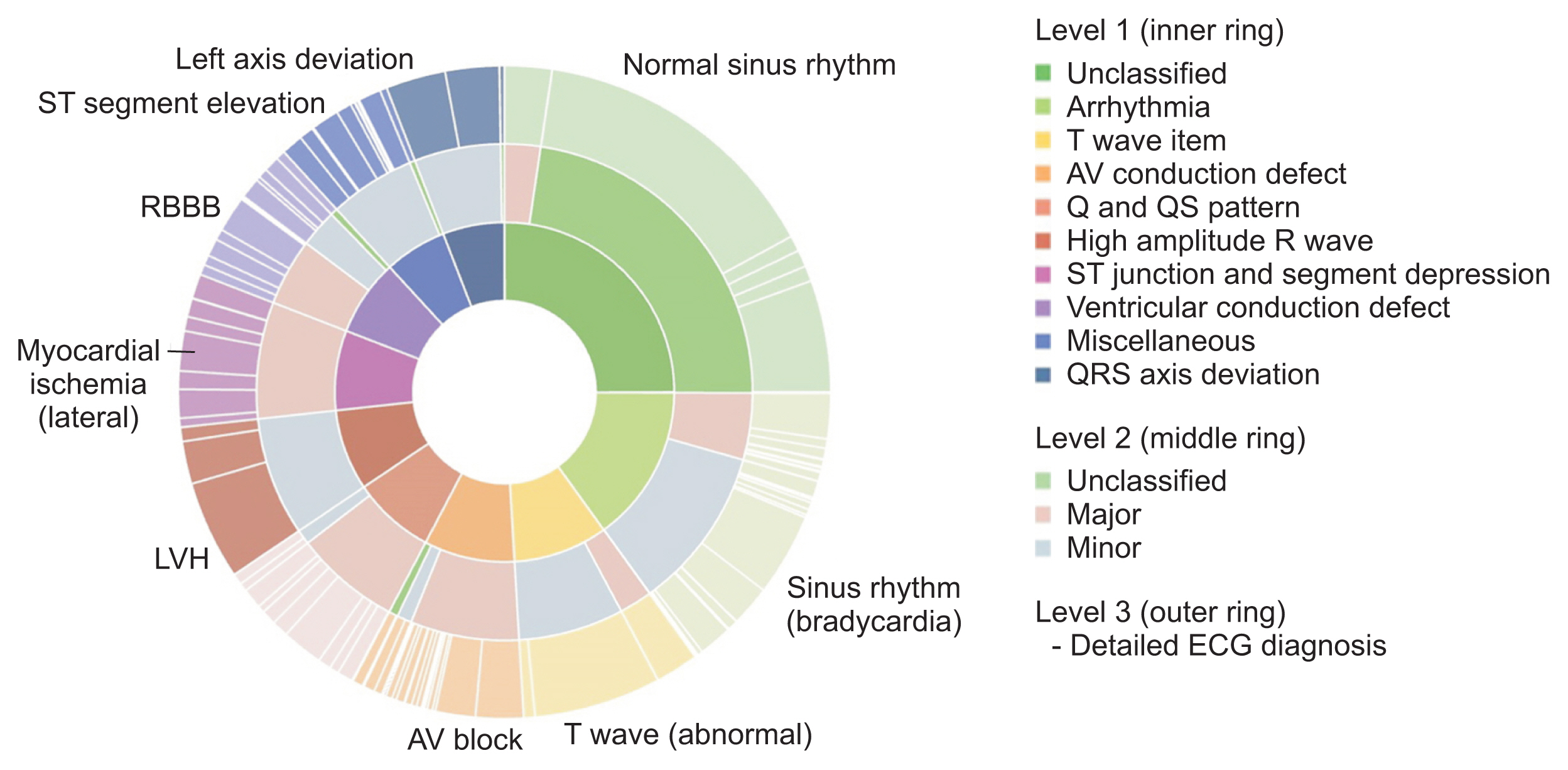
The constructed database was standardized using Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) and Observational Medical Outcomes Partnership–common data model (OMOP-CDM), and data were then categorized into 10 groups based on the Minnesota classification. In addition, to extract high-quality waveforms, poor-quality ECGs were removed, and database bias was minimized by extracting at least 2,000 cases for each group. To check database quality, the difference in baseline displacement according to whether poor ECGs were removed was analyzed, and the usefulness of the database was verified with seven classification models using waveforms.
Results
The standardized KURIAS-ECG database consists of high-quality ECGs from 13,862 patients, with about 20,000 data points, making it possible to obtain more than 2,000 for each Minnesota classification. An artificial intelligence classification model using the data extracted through SNOMED-CT showed an average accuracy of 88.03%.
Conclusions
The KURIAS-ECG database contains standardized ECG data extracted from various machines. The proposed protocol should promote cardiovascular disease research using big data and artificial intelligence.
Figure
Reference
-
References
1. Maron BJ, Friedman RA, Kligfield P, Levine BD, Viskin S, Chaitman BR, et al. Assessment of the 12-lead ECG as a screening test for detection of cardiovascular disease in healthy general populations of young people (12–25 years of age): a scientific statement from the American Heart Association and the American College of Cardiology. Circulation. 2014; 130(15):1303–34. https://doi.org/10.1161/CIR.0000000000000025.
Article2. Hao P, Gao X, Li Z, Zhang J, Wu F, Bai C. Multi-branch fusion network for Myocardial infarction screening from 12-lead ECG images. Comput Methods Programs Biomed. 2020; 184:105286. https://doi.org/10.1016/j.cmpb.2019.105286.
Article3. Rajkumar A, Ganesan M, Lavanya R. Arrhythmia classification on ECG using deep learning. In : Proceedings of 2019 5th International Conference on advanced Computing & Communication Systems (ICACCS); 2019 Mar 15–16; Coimbatore, India. p. 365–9. https://doi.org/10.1109/ICACCS.2019.8728362.
Article4. Liu X, Wang H, Li Z, Qin L. Deep learning in ECG diagnosis: a review. Knowl Based Syst. 2021; 227:107187. https://doi.org/10.1016/j.knosys.2021.107187.
Article5. Hong S, Zhou Y, Shang J, Xiao C, Sun J. Opportunities and challenges of deep learning methods for electrocardiogram data: a systematic review. Comput Biol Med. 2020; 122:103801. https://doi.org/10.1016/j.compbiomed.2020.103801.
Article6. Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PCh, Mark RG, Mietus JE, Moody GB, Peng C-K, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000. 101:e215–e220. http://circ.ahajournals.org/cgi/content/full/101/23/e215.7. Taddei A, Distante G, Emdin M, Pisani P, Moody GB, Zeelenberg C, et al. The European ST-T database: standard for evaluating systems for the analysis of ST-T changes in ambulatory electrocardiography. Eur Heart J. 1992; 13(9):1164–72. https://doi.org/10.1093/oxfordjournals.eurheartj.a060332.
Article8. Jager F, Taddei A, Moody GB, Emdin M, Antolic G, Dorn R, et al. Long-term ST database: a reference for the development and evaluation of automated ischaemia detectors and for the study of the dynamics of myocardial ischaemia. Med Biol Eng Comput. 2003; 41(2):172–82. https://doi.org/10.1007/BF02344885.
Article9. Moody GB, Mark RG. The impact of the MIT-BIH arrhythmia database. IEEE Eng Med Biol Mag. 2001; 20(3):45–50. https://doi.org/10.1109/51.932724.
Article10. Moody GB, Muldrow W, Mark RG. A noise stress test for arrhythmia detectors. Comput Cardiol. 1984; 11(3):381–4.11. Laguna P, Sornmo L. The STAFF III ECG database and its significance for methodological development and evaluation. J Electrocardiol. 2014; 47(4):408–17. https://doi.org/10.1016/j.jelectrocard.2014.04.018.
Article12. Kreiseler D, Bousseljot R. Automatisierte EKG-Auswertung mit Hilfe der EKG-Signaldatenbank CARDIODAT der PTB. Biomed Tech (Berl). 1995; 40(s1):319–20. https://doi.org/10.1515/bmte.1995.40.s1.319.
Article13. Yakushenko E. St Petersburg INCART 12-lead Arrhythmia Database [Internet]. Cambridge (MA): PhysioNet;2008. [cited at 2023 Mar 30]. Available from: https://physionet.org/content/incartdb/1.0.0/.14. Moody GB. The PhysioNet/Computers in Cardiology challenge 2008: T-wave alternans. In : Proceedings of 2008 Computers in Cardiology; 2008 Sep 14–17; Bologna, Italy. p. 505–8. https://doi.org/10.1109/CIC.2008.4749089.
Article15. Kalyakulina AI, Yusipov II, Moskalenko VA, Nikolskiy AV, Kosonogov KA, Osipov GV, et al. LUDB: a new open-access validation tool for electrocardiogram delineation algorithms. IEEE Access. 2020; 8:186181–90. https://doi.org/10.1109/ACCESS.2020.3029211.
Article16. Zheng J, Zhang J, Danioko S, Yao H, Guo H, Rakovski C. A 12-lead electrocardiogram database for arrhythmia research covering more than 10,000 patients. Sci Data. 2020; 7(1):48. https://doi.org/10.1038/s41597-020-0386-x.
Article17. Wagner P, Strodthoff N, Bousseljot RD, Kreiseler D, Lunze FI, Samek W, et al. PTB-XL, a large publicly available electrocardiography dataset. Sci Data. 2020; 7(1):154. https://doi.org/10.1038/s41597-020-0495-6.
Article18. Donnelly K. SNOMED-CT: the advanced terminology and coding system for eHealth. Stud Health Technol Inform. 2006; 121:279–90.19. Lee D, de Keizer N, Lau F, Cornet R. Literature review of SNOMED CT use. J Am Med Inform Assoc. 2014; 21(e1):e11–9. https://doi.org/10.1136/amiajnl-2013-001636.
Article20. Prineas RJ, Crow RS, Zhang ZM. The Minnesota code manual of electrocardiographic findings. New York (NY): Springer Science & Business Media;2009.21. Willems JL, Abreu-Lima C, Arnaud P, van Bemmel JH, Brohet C, Degani R, et al. The diagnostic performance of computer programs for the interpretation of electrocardiograms. N Engl J Med. 1991; 325(25):1767–73. https://doi.org/10.1056/NEJM199112193252503.
Article22. Smulyan H. The computerized ECG: friend and foe. Am J Med. 2019; 132(2):153–60. https://doi.org/10.1016/j.amjmed.2018.08.025.
Article23. Makadia R, Ryan PB. Transforming the premier perspective hospital database into the Observational Medical Outcomes Partnership (OMOP) common data model. EGEMS (Wash DC). 2014; 2(1):1110. https://doi.org/10.13063/2327-9214.1110.
Article24. Altay Y, Kremlev A, Zimenko K, Margun A. The effect of filter parameters on the accuracy of ECG signal measurement. Biomed Eng. 2019; 53(3):176–80. https://doi.org/10.1007/s10527-019-09903-2.
Article25. Sohn J, Yang S, Lee J, Ku Y, Kim HC. Reconstruction of 12-lead electrocardiogram from a three-lead patch-type device using a LSTM network. Sensors (Basel). 2020; 20(11):3278. https://doi.org/10.3390/s20113278.
Article26. Gan F, Ruan G, Mo J. Baseline correction by improved iterative polynomial fitting with automatic threshold. Chemometr Intell Lab Syst. 2006; 82(1–2):59–65. https://doi.org/10.1016/j.chemolab.2005.08.009.
Article27. Baek SJ, Park A, Ahn YJ, Choo J. Baseline correction using asymmetrically reweighted penalized least squares smoothing. Analyst. 2015; 140(1):250–7. https://doi.org/10.1039/c4an01061b.
Article28. Meek S, Morris F; ABC of clinical electrocardiography. Introduction. I-Leads, rate, rhythm, and cardiac axis. BMJ. 2002; 324(7334):415–8. https://doi.org/10.1136/bmj.324.7334.415.
Article29. Ebrahimi Z, Loni M, Daneshtalab M, Gharehbaghi A. A review on deep learning methods for ECG arrhythmia classification. Exp Syst Appl. 2020; 7:100033. https://doi.org/10.1016/j.eswax.2020.100033.
Article30. Yoo H, Chung SH, Lee C-N, Joo HJ. Deep Learning Algorithm of 12-Lead Electrocardiogram for Parkinson Disease Screening. J Parkinsons Dis. 2023. Preprint. 1–12. https://doi.org/10.3233/JPD-223549.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Construction of an Electrocardiogram Database Including 12 Lead Waveforms
- Mass Screening of Heart Disease with 12-Lead Electrocardiography(ECG) in Elementary School Children
- An Automated Fast Healthcare InteroperabilityResources-Based 12-Lead Electrocardiogram MobileAlert System for Suspected Acute Coronary Syndrome
- Association between prehospital recognition of acute myocardial infarction and length of stay in the emergency department
- Resting and Exercise Electrocardiograms in Baseball Players