Brain Tumor Res Treat.
2023 Apr;11(2):86-93. 10.14791/btrt.2023.0011.
Epigenetic and Metabolic Changes in Diffuse Intrinsic Pontine Glioma
- Affiliations
-
- 1Department of New Biology, Daegu Gyeongbuk Institute of Science and Technology (DGIST), Daegu, Korea
- 2New Biology Research Center (NBRC), Daegu Gyeongbuk Institute of Science and Technology (DGIST), Daegu, Korea
- KMID: 2542080
- DOI: http://doi.org/10.14791/btrt.2023.0011
Abstract
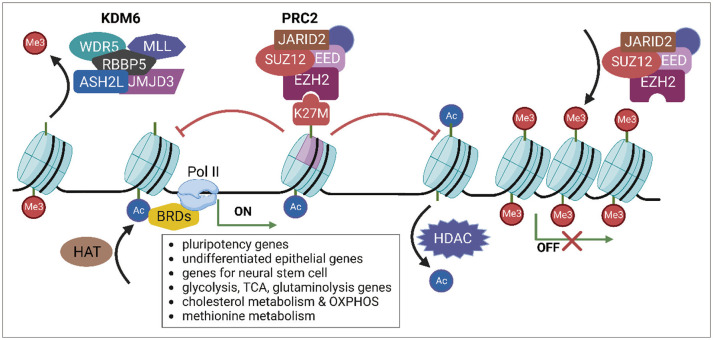
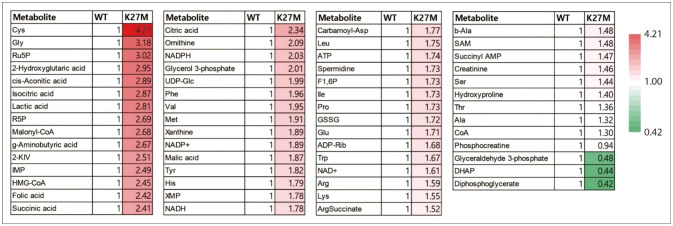
- Diffuse midline glioma (DMG), hitherto known as diffuse intrinsic pontine glioma (DIPG), is a rare and aggressive form of brain cancer that primarily affects children. Although the exact cause of DMG/DIPG is not known, a large proportion of DMG/DIPG tumors harbor mutations in the gene encoding the histone H3 protein, specifically the H3K27M mutation. This mutation decreases the level of H3K27me3, a histone modification that plays a vital role in regulating gene expression through epigenetic regulation. The mutation also alters the function of polycomb repressive complex 2 (PRC2), thereby preventing the repression of genes associated with cancer development. The decrease in H3K27me3 caused by the histone H3 mutation is accompanied by an increase in the level of H3K27ac, a post-translational modification related to active transcription. Dysregulation of histone modification markedly affects gene expression, contributing to cancer development and progression by promoting uncontrolled cell proliferation, tumor growth, and metabolism. DMG/DIPG alters the metabolism of methionine and the tricarboxylic acid cycle, as well as glucose and glutamine uptake. The role of epigenetic and metabolic changes in the development of DMG/DIPG has been studied extensively, and understanding these changes is critical to developing therapies targeting these pathways. Studies are currently underway to identify new therapeutic targets for DMG/DIPG, which may lead to the development of effective treatments for this devastating disease.
Keyword
Figure
Reference
-
1. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016; 131:803–820. PMID: 27157931.2. Hayden E, Holliday H, Lehmann R, Khan A, Tsoli M, Rayner BS, et al. Therapeutic targets in diffuse midline gliomas—an emerging landscape. Cancers (Basel). 2021; 13:6251. PMID: 34944870.3. Damodharan S, Lara-Velazquez M, Williamsen BC, Helgager J, Dey M. Diffuse intrinsic pontine glioma: molecular landscape, evolving treatment strategies and emerging clinical trials. J Pers Med. 2022; 12:840. PMID: 35629262.4. Vitanza NA, Monje M. Diffuse intrinsic pontine glioma: from diagnosis to next-generation clinical trials. Curr Treat Options Neurol. 2019; 21:37. PMID: 31290035.5. Wierzbicki K, Ravi K, Franson A, Bruzek A, Cantor E, Harris M, et al. Targeting and therapeutic monitoring of H3K27M-mutant glioma. Curr Oncol Rep. 2020; 22:19. PMID: 32030483.6. Grimm SA, Chamberlain MC. Brainstem glioma: a review. Curr Neurol Neurosci Rep. 2013; 13:346. PMID: 23512689.7. Jansen MH, Veldhuijzen van Zanten SE, Sanchez Aliaga E, Heymans MW, Warmuth-Metz M, Hargrave D, et al. Survival prediction model of children with diffuse intrinsic pontine glioma based on clinical and radiological criteria. Neuro Oncol. 2014; 17:160–166. PMID: 24903904.8. Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012; 44:251–253. PMID: 22286216.9. Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012; 482:226–231. PMID: 22286061.10. Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012; 22:425–437. PMID: 23079654.11. Kornberg RD, Lorch Y. Primary role of the nucleosome. Mol Cell. 2020; 79:371–375. PMID: 32763226.12. Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012; 124:439–447. PMID: 22661320.13. Castel D, Philippe C, Calmon R, Le Dret L, Truffaux N, Boddaert N, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015; 130:815–827. PMID: 26399631.14. Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 2016; 8:a019521. PMID: 27037415.15. Sievers P, Sill M, Schrimpf D, Stichel D, Reuss DE, Sturm D, et al. A subset of pediatric-type thalamic gliomas share a distinct DNA methylation profile, H3K27me3 loss and frequent alteration of EGFR. Neuro Oncol. 2020; 23:34–43.16. Wen PY, Packer RJ. The 2021 WHO classification of tumors of the central nervous system: clinical implications. Neuro Oncol. 2021; 23:1215–1217. PMID: 34185090.17. Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013; 340:857–861. PMID: 23539183.18. Harutyunyan AS, Krug B, Chen H, Papillon-Cavanagh S, Zeinieh M, De Jay N, et al. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat Commun. 2019; 10:1262. PMID: 30890717.19. Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, et al. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. 2013; 23:558–564. PMID: 23414300.20. Pun M, Pratt D, Nano PR, Joshi PK, Jiang L, Englinger B, et al. Common molecular features of H3K27M DMGs and PFA ependymomas map to hindbrain developmental pathways. Acta Neuropathol Commun. 2023; 11:25. PMID: 36759899.21. Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science. 2018; 360:331–335. PMID: 29674595.22. Lewis NA, Klein RH, Kelly C, Yee J, Knoepfler PS. Histone H3.3 K27M chromatin functions implicate a network of neurodevelopmental factors including ASCL1 and NEUROD1 in DIPG. Epigenetics Chromatin. 2022; 15:18. PMID: 35590427.23. Brien GL, Bressan RB, Monger C, Gannon D, Lagan E, Doherty AM, et al. Simultaneous disruption of PRC2 and enhancer function underlies histone H3.3-K27M oncogenic activity in human hindbrain neural stem cells. Nat Genet. 2021; 53:1221–1232. PMID: 34294917.24. Larson JD, Kasper LH, Paugh BS, Jin H, Wu G, Kwon CH, et al. Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell. 2019; 35:140–155.e7. PMID: 30595505.25. Kfoury-Beaumont N, Prakasam R, Pondugula S, Lagas JS, Matkovich S, Gontarz P, et al. The H3K27M mutation alters stem cell growth, epigenetic regulation, and differentiation potential. BMC Biol. 2022; 20:124. PMID: 35637482.26. Lulla RR, Saratsis AM, Hashizume R. Mutations in chromatin machinery and pediatric high-grade glioma. Sci Adv. 2016; 2:e1501354. PMID: 27034984.27. Laugesen A, Højfeldt JW, Helin K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol Cell. 2019; 74:8–18. PMID: 30951652.28. Harutyunyan AS, Chen H, Lu T, Horth C, Nikbakht H, Krug B, et al. H3K27M in gliomas causes a one-step decrease in H3K27 methylation and reduced spreading within the constraints of H3K36 methylation. Cell Rep. 2020; 33:108390. PMID: 33207202.29. Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA, et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med. 2017; 23:493–500. PMID: 28263307.30. Leszczynska KB, Jayaprakash C, Kaminska B, Mieczkowski J. Emerging advances in combinatorial treatments of epigenetically altered pediatric high-grade H3K27M gliomas. Front Genet. 2021; 12:742561. PMID: 34646308.31. Krug B, De Jay N, Harutyunyan AS, Deshmukh S, Marchione DM, Guilhamon P, et al. Pervasive H3K27 acetylation leads to ERV expression and a therapeutic vulnerability in H3K27M gliomas. Cancer Cell. 2019; 35:782–797.e8. PMID: 31085178.32. Silveira AB, Kasper LH, Fan Y, Jin H, Wu G, Shaw TI, et al. H3.3 K27M depletion increases differentiation and extends latency of diffuse intrinsic pontine glioma growth in vivo. Acta Neuropathol. 2019; 137:637–655. PMID: 30770999.33. Furth N, Algranati D, Dassa B, Beresh O, Fedyuk V, Morris N, et al. H3-K27M-mutant nucleosomes interact with MLL1 to shape the glioma epigenetic landscape. Cell Rep. 2022; 39:110836. PMID: 35584667.34. Harpaz N, Mittelman T, Beresh O, Griess O, Furth N, Salame TM, et al. Single-cell epigenetic analysis reveals principles of chromatin states in H3.3-K27M gliomas. Mol Cell. 2022; 82:2696–2713.e9. PMID: 35716669.35. Gates LA, Foulds CE, O’Malley BW. Histone marks in the ‘driver’s seat’: functional roles in steering the transcription cycle. Trends Biochem Sci. 2017; 42:977–989. PMID: 29122461.36. Miyai M, Kanayama T, Hyodo F, Kinoshita T, Ishihara T, Okada H, et al. Glucose transporter Glut1 controls diffuse invasion phenotype with perineuronal satellitosis in diffuse glioma microenvironment. Neurooncol Adv. 2020; 3:vdaa150. PMID: 33506198.37. Autry AW, Hashizume R, James CD, Larson PEZ, Vigneron DB, Park I. Measuring tumor metabolism in pediatric diffuse intrinsic pontine glioma using hyperpolarized carbon-13 MR metabolic imaging. Contrast Media Mol Imaging. 2018; 2018:3215658. PMID: 30174560.38. Mbah NE, Myers AL, Chung C, Thompson JK, Hong HS, Sajjakulnukit P. . Therapeutic targeting of differentiation state-dependent metabolic vulnerabilities in DIPG. bioRxiv [Preprint]. 2022; cited 2023 Mar 24. DOI: 10.1101/2022.03.01.482555.39. Chung C, Sweha SR, Pratt D, Tamrazi B, Panwalkar P, Banda A, et al. Integrated metabolic and epigenomic reprograming by H3K27M mutations in diffuse intrinsic pontine gliomas. Cancer Cell. 2020; 38:334–349.e9. PMID: 32795401.40. Ijare O, Manalo J, Sharpe M, Baskin D, Pichumani K. TAMI-80. Cellular metabolism in diffuse intrinsic pontine glioma. Neuro Oncol. 2021; 23(Suppl 6):vi215.41. Golbourn BJ, Halbert ME, Halligan K, Varadharajan S, Krug B, Mbah NE, et al. Loss of MAT2A compromises methionine metabolism and represents a vulnerability in H3K27M mutant glioma by modulating the epigenome. Nat Cancer. 2022; 3:629–648. PMID: 35422502.42. Parsels LA, Wahl DR, Koschmann C, Morgan MA, Zhang Q. Developing H3K27M mutant selective radiosensitization strategies in diffuse intrinsic pontine glioma. Neoplasia. 2023; 37:100881. PMID: 36724689.43. Franson A, Koschmann C. Enhancing GD2 CAR T-cell therapy with IGF-1R blockade: are DIPG CAR T cells ready for combinatorial therapy? Neuro Oncol. 2022; 24:1164–1165. PMID: 35323961.44. Messinger D, Harris MK, Cummings JR, Thomas C, Yang T, Sweha SR, et al. Therapeutic targeting of prenatal pontine ID1 signaling in diffuse midline glioma. Neuro Oncol. 2023; 25:54–67. PMID: 35605606.45. Gardner SL, Tarapore RS, Allen J, McGovern SL, Zaky W, Odia Y, et al. Phase I dose escalation and expansion trial of single agent ONC201 in pediatric diffuse midline gliomas following radiotherapy. Neurooncol Adv. 2022; 4:vdac143. PMID: 36382108.46. Mohammad F, Weissmann S, Leblanc B, Pandey DP, Højfeldt JW, Comet I, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017; 23:483–492. PMID: 28263309.47. Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014; 20:1394–1396. PMID: 25401693.48. Katagi H, Louis N, Unruh D, Sasaki T, He X, Zhang A, et al. Radiosensitization by histone H3 demethylase inhibition in diffuse intrinsic pontine glioma. Clin Cancer Res. 2019; 25:5572–5583. PMID: 31227500.49. Nikolaev A, Fiveash JB, Yang ES. Combined targeting of mutant p53 and Jumonji family histone demethylase augments therapeutic efficacy of radiation in H3K27M DIPG. Int J Mol Sci. 2020; 21:490. PMID: 31940975.50. Nagaraja S, Vitanza NA, Woo PJ, Taylor KR, Liu F, Zhang L, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell. 2017; 31:635–652.e6. PMID: 28434841.51. Lin GL, Wilson KM, Ceribelli M, Stanton BZ, Woo PJ, Kreimer S, et al. Therapeutic strategies for diffuse midline glioma from high-throughput combination drug screening. Sci Transl Med. 2019; 11:eaaw0064. PMID: 31748226.52. Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015; 21:555–559. PMID: 25939062.53. Harris W. A case of pontine glioma, with special reference to the paths of gustatory sensation. Proc R Soc Med. 1926; 19:1–5.54. Zhao X, Li D, Qiao Z, Wang K, Chen Q, Pan C, et al. 11C-methionine PET imaging characteristics in children with diffuse intrinsic pontine gliomas and relationship to survival and H3 K27M mutation status. Eur J Nucl Med Mol Imaging. 2023; 01. 26. DOI: 10.1007/s00259-022-06105-z. [Epub].55. Pal S, Kaplan JP, Nguyen H, Stopka SA, Savani MR, Regan MS, et al. A druggable addiction to de novo pyrimidine biosynthesis in diffuse midline glioma. Cancer Cell. 2022; 40:957–972.e10. PMID: 35985342.56. Mersich I, Dasgupta B. DIPG-03. Therapeutic targeting of purine biosynthesis in DIPG. Neuro Oncol. 2022; 24(Suppl 1):i17.57. Zhang Y, Zhou L, Safran H, Borsuk R, Lulla R, Tapinos N, et al. EZH2i EPZ-6438 and HDACi vorinostat synergize with ONC201/TIC10 to activate integrated stress response, DR5, reduce H3K27 methylation, ClpX and promote apoptosis of multiple tumor types including DIPG. Neoplasia. 2021; 23:792–810. PMID: 34246076.58. Su JM, Murray JC, McNall-Knapp RY, Bowers DC, Shah S, Adesina AM, et al. A phase 2 study of valproic acid and radiation, followed by maintenance valproic acid and bevacizumab in children with newly diagnosed diffuse intrinsic pontine glioma or high-grade glioma. Pediatr Blood Cancer. 2020; 67:e28283. PMID: 32285998.59. Su JM, Li XN, Thompson P, Ou CN, Ingle AM, Russell H, et al. Phase 1 study of valproic acid in pediatric patients with refractory solid or CNS tumors: a children’s oncology group report. Clin Cancer Res. 2011; 17:589–597. PMID: 21115653.60. Wang J, Huang TY, Hou Y, Bartom E, Lu X, Shilatifard A, et al. Epigenomic landscape and 3D genome structure in pediatric high-grade glioma. Sci Adv. 2021; 7:eabg4126. PMID: 34078608.61. Chi AS, Stafford JM, Sen N, Possemato R, Placantonakis D, Hidalgo ET, et al. EXTH-42. H3 K27M mutant gliomas are selectively killed by ONC201, a small molecule inhibitor of dopamine receptor D2. Neuro Oncol. 2017; 19(suppl 6):vi81.62. Lauing K, Lulla R, Lenzen A, Zhai L, Hashizume R, Fangusaro JR, et al. IMMU-35. Targeting IDO1 in human pediatric brain cancer. Neuro Oncol. 2017; 19(Suppl 6):vi120.63. Lauing KL, Lulla RR, Zhai L, Hashizume R, Fangusaro J, Wainwright DA. IMMU-21. Characterizing IDO1 and its therapeutic potential in pediatric central nervous system tumors. Neuro-Oncol. 2017; 19(Suppl 4):iv32.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Radiotherapy for Diffuse Intrinsic Pontine Glioma: Insufficient but Indispensable
- Spontaneous Regression of Glioma–Mimicking Brainstem Lesion in a Child: A Case Report
- Diffuse Intrinsic Pontine Glioma : Clinical Features, Molecular Genetics, and Novel Targeted Therapeutics
- Malignant Brain Tumours in Children : Present and Future Perspectives
- Diffuse Midline Gliomas Harboring the H3 K27M-Mutation in the Bilateral Thalamus and Midbrain: A Case Report and a Review of the Literature