J Korean Med Sci.
2023 Apr;38(15):e120. 10.3346/jkms.2023.38.e120.
Gut Microbiota Dysbiosis Correlates With Long COVID-19 at One-Year After Discharge
- Affiliations
-
- 1Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- KMID: 2541558
- DOI: http://doi.org/10.3346/jkms.2023.38.e120
Abstract
- Background
Long coronavirus disease 2019 (COVID-19) in recovered patients (RPs) is gradually recognized by more people. However, how long it will last and the underlining mechanism remains unclear.
Methods
We conducted a prospective follow-up study to evaluate the long-term symptoms and clinical indices of RPs at one-year after discharge from Union Hospital, Wuhan, China between December 2020 to May 2021. We also performed the 16S rRNA sequencing of stool samples from RPs and healthy controls (HCs) and analyzed the correlation between the gut microbiota and long COVID-19.
Results
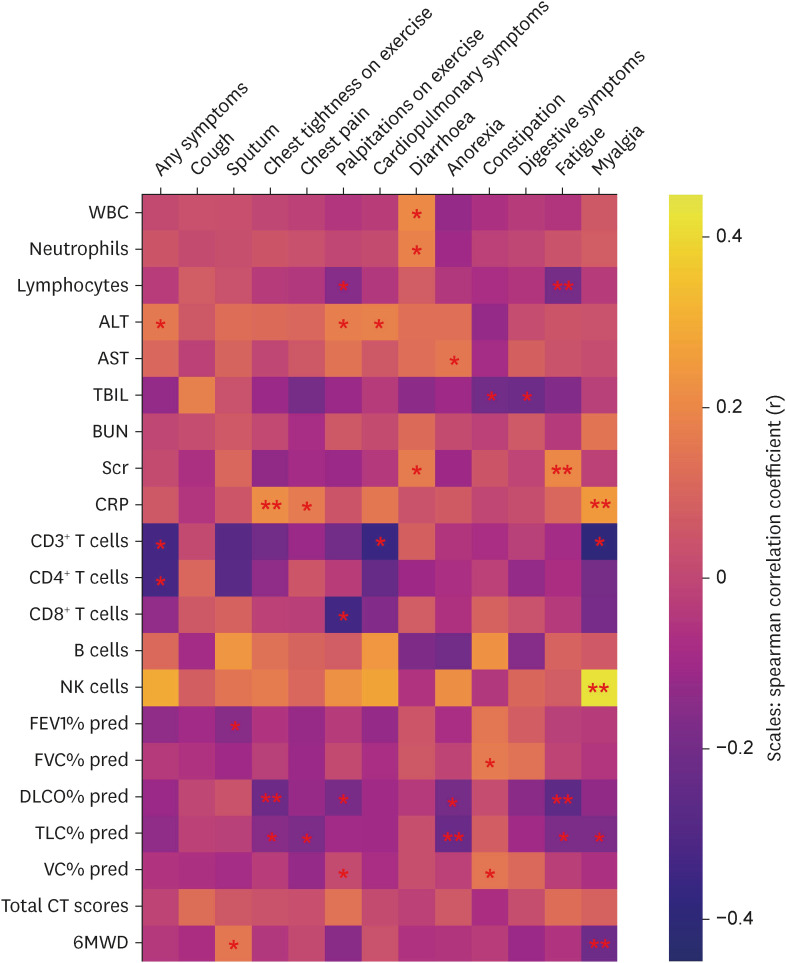
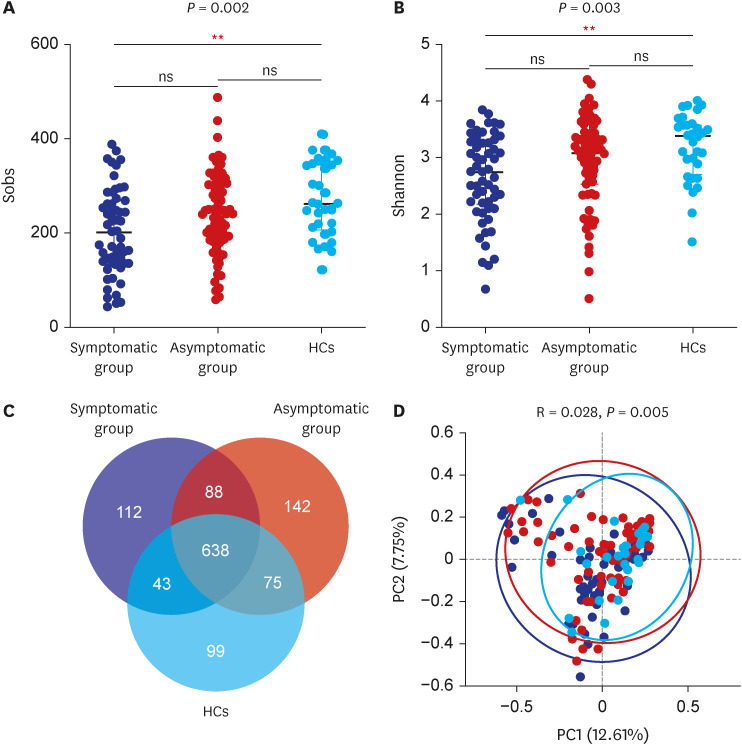
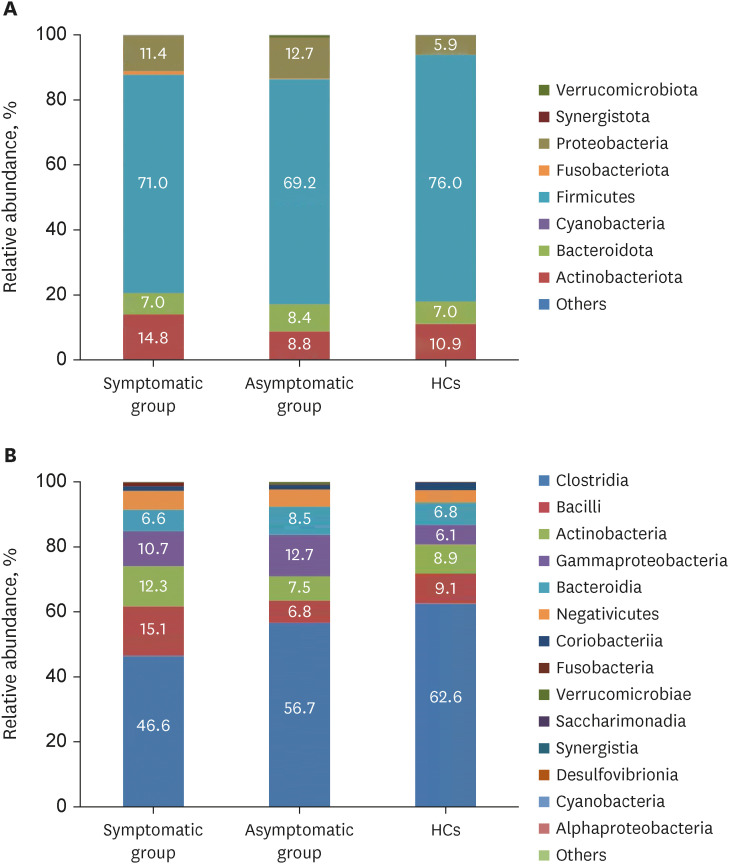
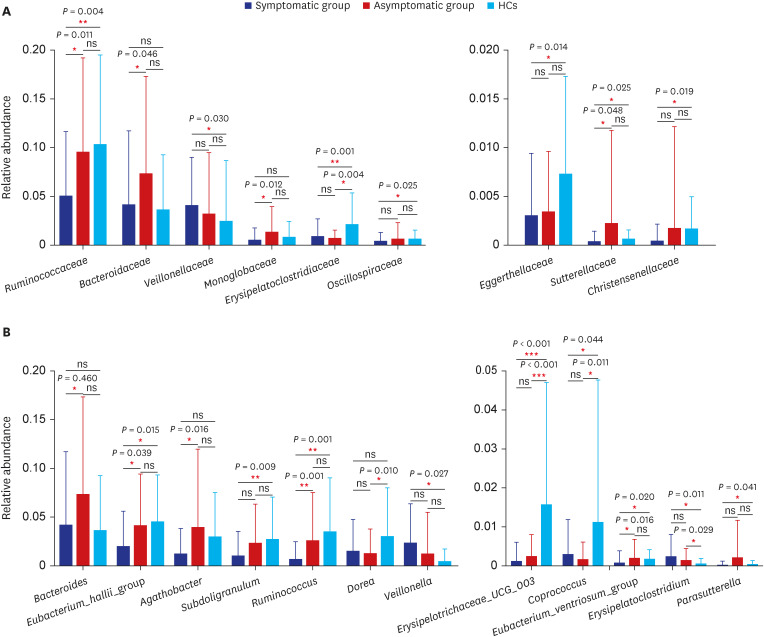
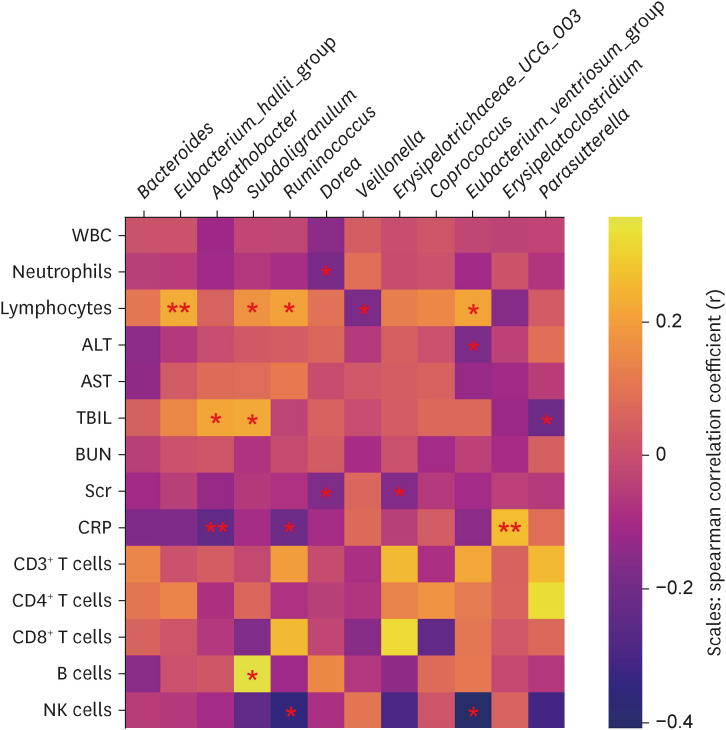
In total, 187 RPs were enrolled, among them, 84 (44.9%) RPs reported long COVID-19 symptoms at one-year after discharge. The most common long-term symptoms were cardiopulmonary symptoms, including chest tightness after activity (39/187, 20.9%), palpitations on exercise (27/187, 14.4%), sputum (21/187, 11.2%), cough (15/187, 8.0%) and chest pain (13/187, 7.0%), followed by systemic symptoms including fatigue (34/187, 18.2%) and myalgia (20/187, 10.7%), and digestive symptoms including constipation (14/187, 7.5%), anorexia (13/187, 7.0%), and diarrhea (8/187, 4.3%). Sixty-six (35.9%) RPs presented either anxiety or depression (42/187 [22.8%] and 53/187 [28.8%] respectively), and the proportion of anxiety or depression in the long symptomatic group was significantly higher than that in the asymptomatic group (41/187 [50.6%] vs. 25/187 [24.3%]). Compared with the asymptomatic group, scores of all nine 36-Item Short Form General Health Survey domains were lower in the symptomatic group (all P < 0.05). One hundred thirty RPs and 32 HCs (non-severe acute respiratory syndrome coronavirus 2 infected subjects) performed fecal sample sequencing. Compared with HCs, symptomatic RPs had obvious gut microbiota dysbiosis including significantly reduced bacterial diversities and lower relative abundance of short-chain fatty acids (SCFAs)-producing salutary symbionts such as Eubacterium_hallii_group, Subdoligranulum, Ruminococcus, Dorea, Coprococcus, and Eubacterium_ventriosum_group. Meanwhile, the relative abundance of Eubacterium_hallii_group, Subdoligranulum, and Ruminococcus showed decreasing tendencies between HCs, the asymptomatic group, and the symptomatic group.
Conclusion
This study demonstrated the presence of long COVID-19 which correlates with gut microbiota dysbiosis in RPs at one-year after discharge, indicating gut microbiota may play an important role in long COVID-19.
Figure
Reference
-
1. Lerum TV, Aaløkken TM, Brønstad E, Aarli B, Ikdahl E, Lund KM, et al. Dyspnoea, lung function and CT findings 3 months after hospital admission for COVID-19. Eur Respir J. 2021; 57(4):2003448. PMID: 33303540.2. Huang C, Huang L, Wang Y, Li X, Ren L, Gu X, et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021; 397(10270):220–232. PMID: 33428867.3. Wu X, Liu X, Zhou Y, Yu H, Li R, Zhan Q, et al. 3-month, 6-month, 9-month, and 12-month respiratory outcomes in patients following COVID-19-related hospitalisation: a prospective study. Lancet Respir Med. 2021; 9(7):747–754. PMID: 33964245.4. Yelin D, Margalit I, Yahav D, Runold M, Bruchfeld J. Long COVID-19-it’s not over until? Clin Microbiol Infect. 2021; 27(4):506–508. PMID: 33316400.5. Shah W, Hillman T, Playford ED, Hishmeh L. Managing the long term effects of covid-19: summary of NICE, SIGN, and RCGP rapid guideline. BMJ. 2021; 372:n136. PMID: 33483331.6. World Health Organization. A clinical case definition of post COVID-19 condition by a Delphi consensus, 6 October 2021. Updated 2021. Accessed October 6, 2021. https://www.who.int/publications/i/item/WHO-2019-nCoV-Post_COVID-19_condition-Clinical_case_definition-2021.1 .7. Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017; 15(1):55–63. PMID: 27694885.8. Pflughoeft KJ, Versalovic J. Human microbiome in health and disease. Annu Rev Pathol. 2012; 7(1):99–122. PMID: 21910623.9. Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021; 70(4):698–706. PMID: 33431578.10. Chen Y, Gu S, Chen Y, Lu H, Shi D, Guo J, et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut. 2022; 71(1):222–225. PMID: 33833065.11. Ren Z, Wang H, Cui G, Lu H, Wang L, Luo H, et al. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut. 2021; 70(7):1253–1265. PMID: 33789966.12. Zhou Y, Zhang J, Zhang D, Ma WL, Wang X. Linking the gut microbiota to persistent symptoms in survivors of COVID-19 after discharge. J Microbiol. 2021; 59(10):941–948. PMID: 34382150.13. Zhou Y, Shi X, Fu W, Xiang F, He X, Yang B, et al. Gut microbiota dysbiosis correlates with abnormal immune response in moderate COVID-19 patients with fever. J Inflamm Res. 2021; 14:2619–2631. PMID: 34168484.14. World Health Organization. COVID-19 clinical management: living guidance, 25 January 2021. Updated 2021. Accessed January 25, 2021. https://apps.who.int/iris/handle/10665/338882 .15. Liang L, Yang B, Jiang N, Fu W, He X, Zhou Y, et al. Three-month follow-up study of survivors of coronavirus disease 2019 after discharge. J Korean Med Sci. 2020; 35(47):e418. PMID: 33289374.16. Wang X, Zhou Q, He Y, Liu L, Ma X, Wei X, et al. Nosocomial outbreak of COVID-19 pneumonia in Wuhan, China. Eur Respir J. 2020; 55(6):2000544. PMID: 32366488.17. Antonelli M, Pujol JC, Spector TD, Ourselin S, Steves CJ. Risk of long COVID associated with delta versus omicron variants of SARS-CoV-2. Lancet. 2022; 399(10343):2263–2264. PMID: 35717982.18. Tansey CM, Louie M, Loeb M, Gold WL, Muller MP, de Jager J, et al. One-year outcomes and health care utilization in survivors of severe acute respiratory syndrome. Arch Intern Med. 2007; 167(12):1312–1320. PMID: 17592106.19. Ong KC, Ng AW, Lee LS, Kaw G, Kwek SK, Leow MK, et al. 1-year pulmonary function and health status in survivors of severe acute respiratory syndrome. Chest. 2005; 128(3):1393–1400. PMID: 16162734.20. Hui DS, Wong KT, Ko FW, Tam LS, Chan DP, Woo J, et al. The 1-year impact of severe acute respiratory syndrome on pulmonary function, exercise capacity, and quality of life in a cohort of survivors. Chest. 2005; 128(4):2247–2261. PMID: 16236881.21. Lam MH, Wing YK, Yu MW, Leung CM, Ma RC, Kong AP, et al. Mental morbidities and chronic fatigue in severe acute respiratory syndrome survivors: long-term follow-up. Arch Intern Med. 2009; 169(22):2142–2147. PMID: 20008700.22. Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin Infect Dis. 2020; 71(10):2669–2678. PMID: 32497191.23. Zuo T, Zhang F, Lui GC, Yeoh YK, Li AY, Zhan H, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020; 159(3):944–955.e8. PMID: 32442562.24. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012; 336(6086):1268–1273. PMID: 22674334.25. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021; 19(1):55–71. PMID: 32887946.26. McAleer JP, Kolls JK. Contributions of the intestinal microbiome in lung immunity. Eur J Immunol. 2018; 48(1):39–49. PMID: 28776643.27. Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieërs G, Guery B, et al. The gut-lung axis in health and respiratory diseases: a place for inter-organ and inter-kingdom crosstalks. Front Cell Infect Microbiol. 2020; 10:9. PMID: 32140452.28. Li S, Hua D, Wang Q, Yang L, Wang X, Luo A, et al. The role of bacteria and its derived metabolites in chronic pain and depression: recent findings and research progress. Int J Neuropsychopharmacol. 2020; 23(1):26–41. PMID: 31760425.29. Hu S, Li A, Huang T, Lai J, Li J, Sublette ME, et al. Gut microbiota changes in patients with bipolar depression. Adv Sci (Weinh). 2019; 6(14):1900752. PMID: 31380217.30. Järbrink-Sehgal E, Andreasson A. The gut microbiota and mental health in adults. Curr Opin Neurobiol. 2020; 62:102–114. PMID: 32163822.31. Zhang J, Guo Z, Xue Z, Sun Z, Zhang M, Wang L, et al. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J. 2015; 9(9):1979–1990. PMID: 25647347.32. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014; 12(10):661–672. PMID: 25198138.33. Holmstrøm K, Collins MD, Møller T, Falsen E, Lawson PA. Subdoligranulum variabile gen. nov., sp. nov. from human feces. Anaerobe. 2004; 10(3):197–203. PMID: 16701519.34. Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016; 167(4):1125–1136.e8. PMID: 27814509.35. Ratajczak W, Rył A, Mizerski A, Walczakiewicz K, Sipak O, Laszczyńska M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim Pol. 2019; 66(1):1–12. PMID: 30831575.36. Sublette ME, Cheung S, Lieberman E, Hu S, Mann JJ, Uhlemann AC, et al. Bipolar disorder and the gut microbiome: a systematic review. Bipolar Disord. 2021; 23(6):544–564. PMID: 33512753.37. Qin N, Zheng B, Yao J, Guo L, Zuo J, Wu L, et al. Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci Rep. 2015; 5(1):14771. PMID: 26490635.38. Sencio V, Barthelemy A, Tavares LP, Machado MG, Soulard D, Cuinat C, et al. Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Reports. 2020; 30(9):2934–2947.e6. PMID: 32130898.39. Tian X, Hellman J, Horswill AR, Crosby HA, Francis KP, Prakash A. Elevated gut microbiome-derived propionate levels are associated with reduced sterile lung inflammation and bacterial immunity in mice. Front Microbiol. 2019; 10:159. PMID: 30891007.40. Vaughan A, Frazer ZA, Hansbro PM, Yang IA. COPD and the gut-lung axis: the therapeutic potential of fibre. J Thorac Dis. 2019; 11(Suppl 17):S2173–S2180. PMID: 31737344.41. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014; 20(2):159–166. PMID: 24390308.42. Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011; 108(13):5354–5359. PMID: 21402903.43. Trompette A, Gollwitzer ES, Pattaroni C, Lopez-Mejia IC, Riva E, Pernot J, et al. Dietary fiber confers protection against flu by shaping Ly6c- patrolling monocyte hematopoiesis and CD8+ T cell metabolism. Immunity. 2018; 48(5):992–1005.e8. PMID: 29768180.44. Wu T, Li H, Su C, Xu F, Yang G, Sun K, et al. Microbiota-derived short-chain fatty acids promote LAMTOR2-mediated immune responses in macrophages. mSystems. 2020; 5(6):e00587-20.45. Wu T, Xu F, Su C, Li H, Lv N, Liu Y, et al. Alterations in the gut microbiome and cecal metabolome during Klebsiella pneumoniae-induced pneumosepsis. Front Immunol. 2020; 11:1331. PMID: 32849494.46. Chen D, Qiu YB, Gao ZQ, Wu YX, Wan BB, Liu G, et al. Sodium propionate attenuates the lipopolysaccharide-induced epithelial-mesenchymal transition via the PI3K/Akt/mTOR signaling pathway. J Agric Food Chem. 2020; 68(24):6554–6563. PMID: 32452677.47. Yamawaki Y, Fuchikami M, Morinobu S, Segawa M, Matsumoto T, Yamawaki S. Antidepressant-like effect of sodium butyrate (HDAC inhibitor) and its molecular mechanism of action in the rat hippocampus. World J Biol Psychiatry. 2012; 13(6):458–467. PMID: 21812623.48. Wei Y, Melas PA, Wegener G, Mathé AA, Lavebratt C. Antidepressant-like effect of sodium butyrate is associated with an increase in TET1 and in 5-hydroxymethylation levels in the Bdnf gene. Int J Neuropsychopharmacol. 2014; 18(2):pyu032. PMID: 25618518.