Cancer Res Treat.
2023 Apr;55(2):671-683. 10.4143/crt.2022.251.
Comprehensive Molecular Characterization of Soft Tissue Sarcoma for Prediction of Pazopanib-Based Treatment Response
- Affiliations
-
- 1Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 2Department of Biomedical Convergence Science and Technology, Kyungpook National University, Daegu, Korea
- 3Cell and Matrix Research Institute, Kyungpook National University, Daegu, Korea
- 4Division of Medical Oncology, Department of Internal Medicine, Yonsei University College of Medicine, Seoul, Korea
- 5Department of Radiology, Yonsei University College of Medicine, Seoul, Korea
- 6Department of Orthopedic Surgery, Yonsei University College of Medicine, Seoul, Korea
- 7Institute for Human Tissue Restoration, Department of Plastic & Reconstructive Surgery, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 8Department of Pathology, Yonsei University College of Medicine, Seoul, Korea
- 9Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 10Department of Hematology-Oncology, Ajou University School of Medicine, Suwon, Korea
- KMID: 2541254
- DOI: http://doi.org/10.4143/crt.2022.251
Abstract
- Purpose
Even though pazopanib, a multitargeted tyrosine kinase inhibitor, has been approved for refractory soft tissue sarcoma (STS), little is known about the molecular determinants of the response to pazopanib. We performed integrative molecular characterization to identify potential predictors of pazopanib efficacy.
Materials and Methods
We obtained fresh pre-treatment tumor tissue from 35 patients with advanced STS receiving pazopanib-based treatment. Among those, 18 (51.4%) received pazopanib monotherapy, and the remaining 17 (48.6%) received pazopanib in combination with durvalumab, programmed death-ligand 1 blockade. Whole-exome and transcriptome sequencing were performed for each tumor and patient germline DNA.
Results
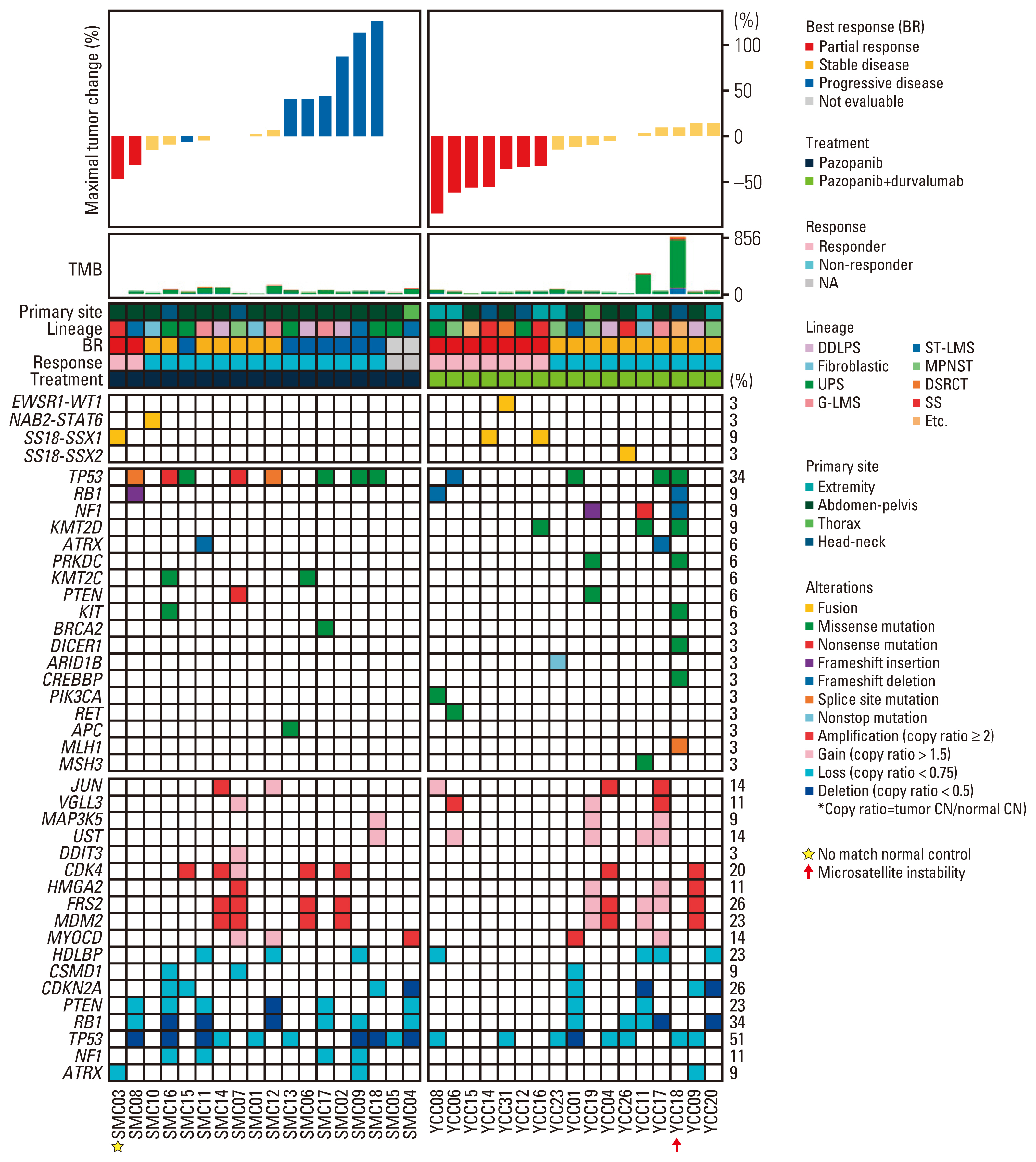
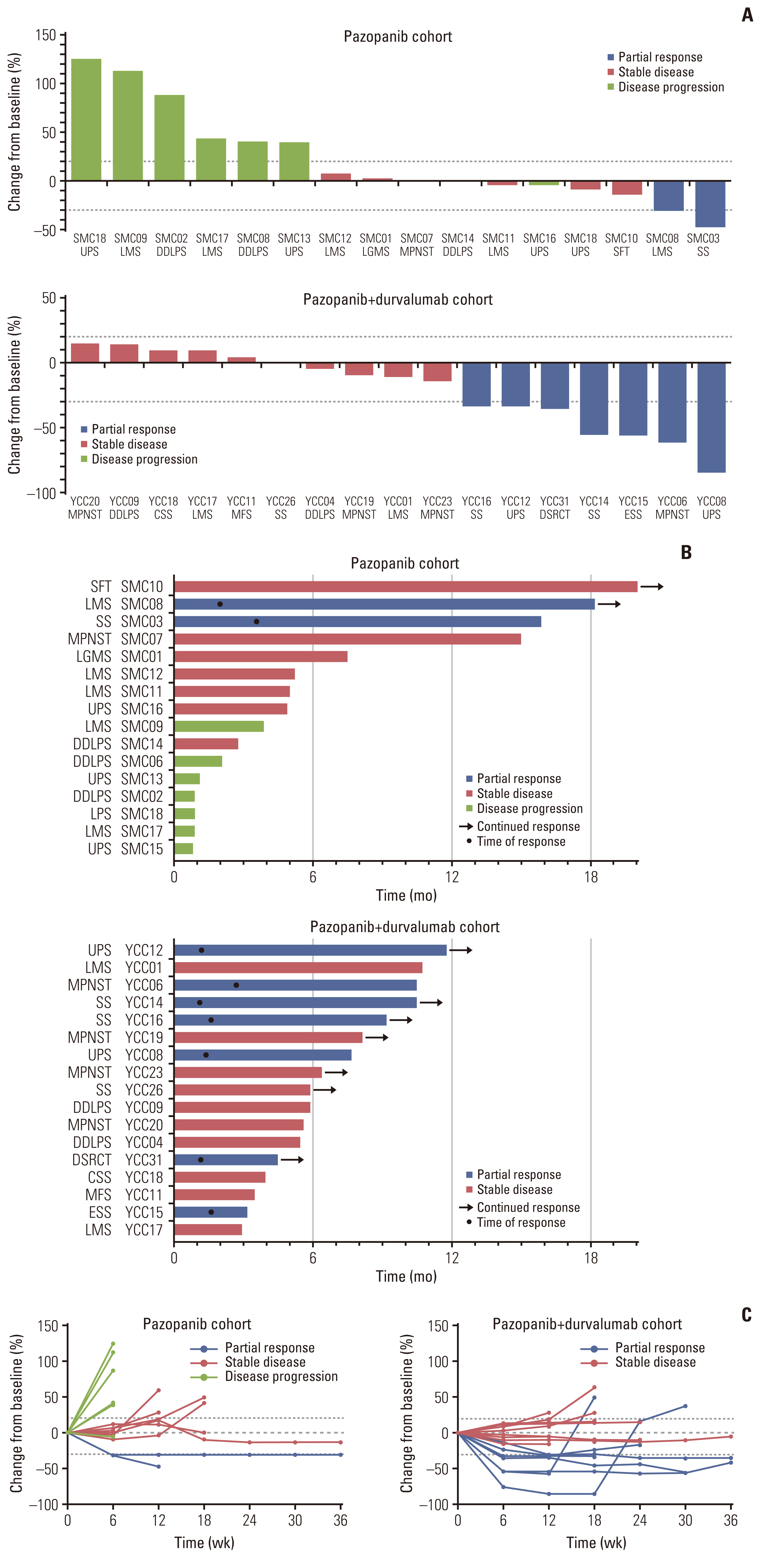
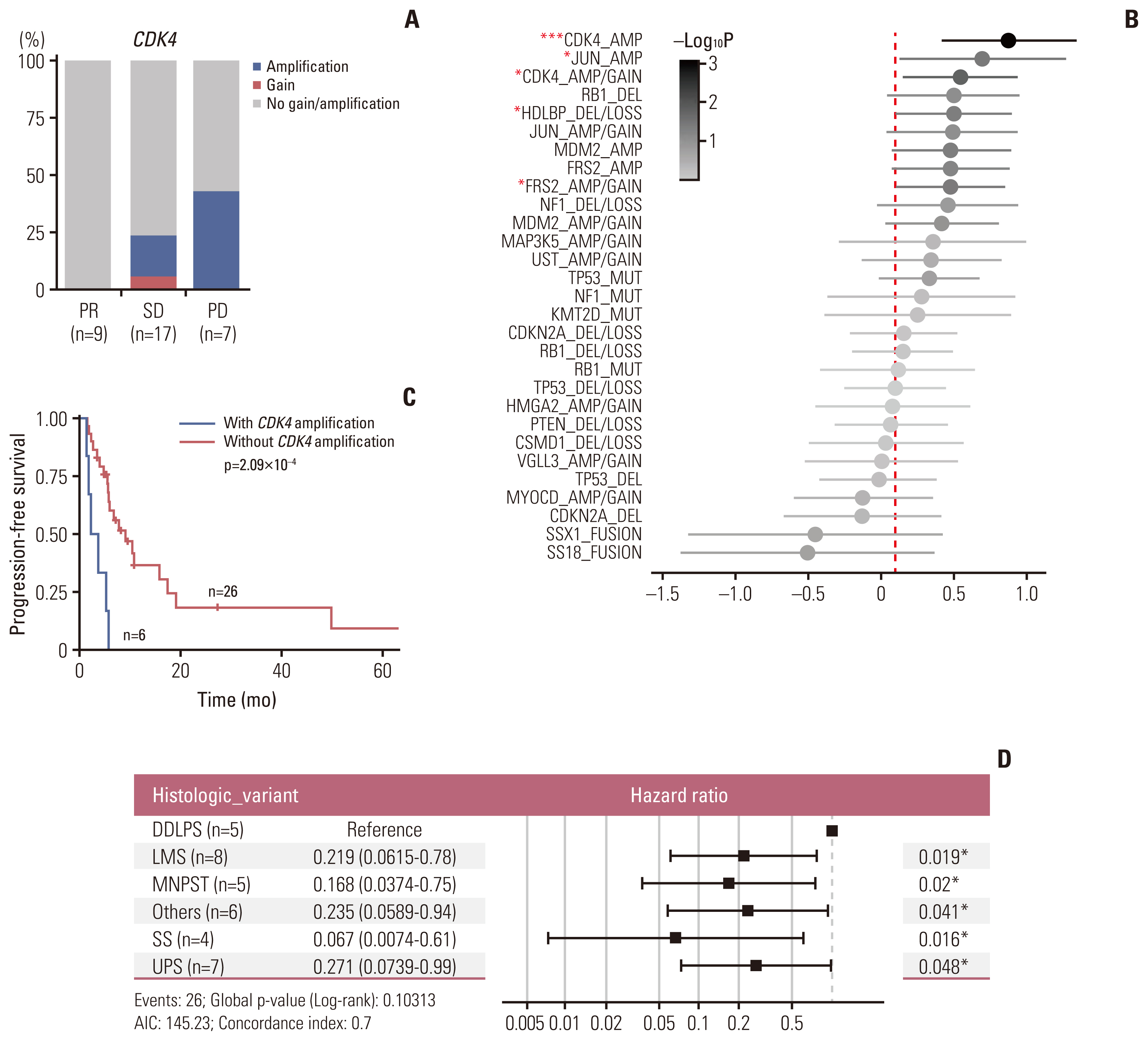
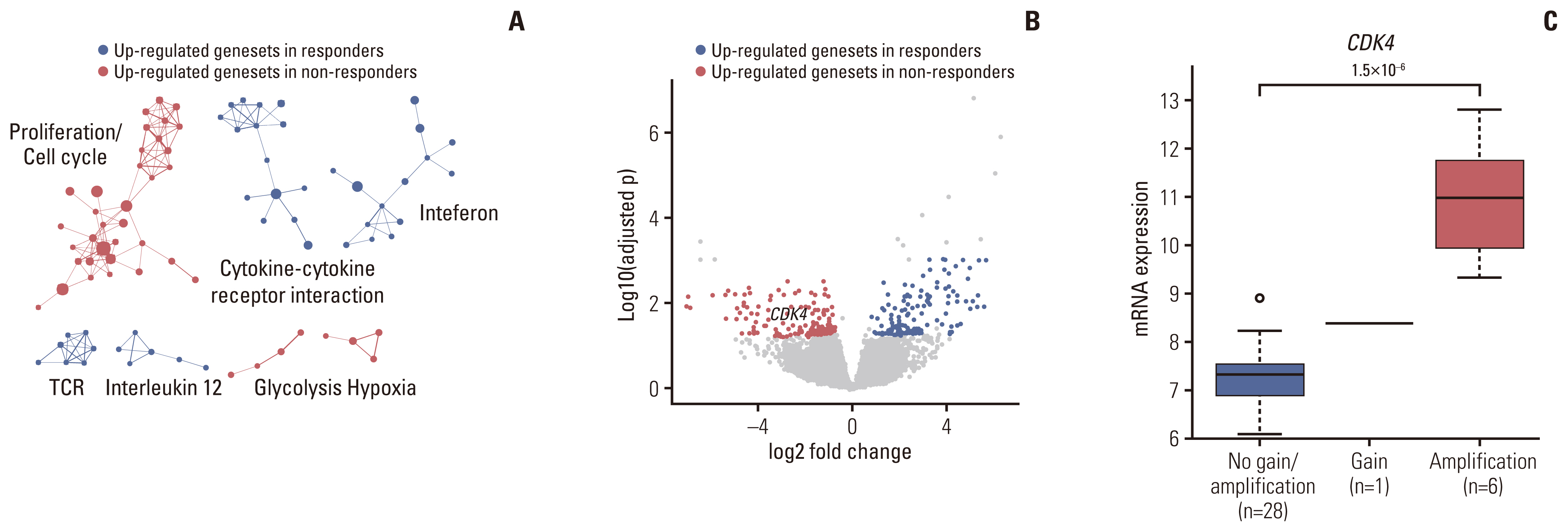
Of the 35 patients receiving pazopanib-based treatment, nine achieved a partial response (PR), resulting in an objective response rate (ORR) of 27.3%, and the median progression-free survival (PFS) was 6.0 months. Patients with CDK4 amplification (copy ratio tumor to normal > 2) exhibited shorter PFS (3.7 vs. 7.9 months, p=2.09×10–4) and a poorer response (ORR; 0% vs. 33.3%) compared to those without a gene amplification (copy ratio ≤ 2). Moreover, non-responders demonstrated transcriptional activation of CDK4 via DNA amplification, resulting in cell cycle activation. In the durvalumab combination cohort, seven of the 17 patients (41.2%) achieved a PR, and gene expression analysis revealed that durvalumab responders exhibited high immune/stromal cell infiltration, mainly comprising natural killer cells, compared to non-responders as well as increased expression of CD19, a B-cell marker.
Conclusion
Despite the limitation of heterogeneity in the study population and treatment, we identified possible molecular predictors of pazopanib efficacy that can be employed in future clinical trials aimed at evaluating therapeutic strategies.
Keyword
Figure
Reference
-
References
1. Hensley ML, Maki R, Venkatraman E, Geller G, Lovegren M, Aghajanian C, et al. Gemcitabine and docetaxel in patients with unresectable leiomyosarcoma: results of a phase II trial. J Clin Oncol. 2002; 20:2824–31.
Article2. Judson I, Verweij J, Gelderblom H, Hartmann JT, Schoffski P, Blay JY, et al. Doxorubicin alone versus intensified doxorubicin plus ifosfamide for first-line treatment of advanced or metastatic soft-tissue sarcoma: a randomised controlled phase 3 trial. Lancet Oncol. 2014; 15:415–23.
Article3. Italiano A, Mathoulin-Pelissier S, Cesne AL, Terrier P, Bonvalot S, Collin F, et al. Trends in survival for patients with metastatic soft-tissue sarcoma. Cancer. 2011; 117:1049–54.
Article4. Sleijfer S, Ray-Coquard I, Papai Z, Le Cesne A, Scurr M, Schoffski P, et al. Pazopanib, a multikinase angiogenesis inhibitor, in patients with relapsed or refractory advanced soft tissue sarcoma: a phase II study from the European organisation for research and treatment of cancer-soft tissue and bone sarcoma group (EORTC study 62043). J Clin Oncol. 2009; 27:3126–32.
Article5. van der Graaf WT, Blay JY, Chawla SP, Kim DW, Bui-Nguyen B, Casali PG, et al. Pazopanib for metastatic soft-tissue sarcoma (PALETTE): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet. 2012; 379:1879–86.
Article6. Tawbi HA, Burgess M, Bolejack V, Van Tine BA, Schuetze SM, Hu J, et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): a multicentre, two-cohort, single-arm, open-label, phase 2 trial. Lancet Oncol. 2017; 18:1493–501.
Article7. Feng X, Pleasance E, Zhao EY, Ng T, Grewal JK, Mohammad N, et al. Therapeutic implication of genomic landscape of adult metastatic sarcoma. JCO Precis Oncol. 2019; 3:1–25.
Article8. Baker SJ, Reddy EP. CDK4: a key player in the cell cycle, development, and cancer. Genes Cancer. 2012; 3:658–69.
Article9. Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016; 17:218.10. Sleijfer S, Gorlia T, Lamers C, Burger H, Blay JY, Le Cesne A, et al. Cytokine and angiogenic factors associated with efficacy and toxicity of pazopanib in advanced soft-tissue sarcoma: an EORTC-STBSG study. Br J Cancer. 2012; 107:639–45.
Article11. Kobayashi H, Okuma T, Oka H, Hirai T, Ohki T, Ikegami M, et al. Neutrophil-to-lymphocyte ratio after pazopanib treatment predicts response in patients with advanced soft-tissue sarcoma. Int J Clin Oncol. 2018; 23:368–74.
Article12. Cancer Genome Atlas Research Network. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017; 171:950–65.13. Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010; 42:715–21.
Article14. Nakamura T, Matsumine A, Kawai A, Araki N, Goto T, Yonemoto T, et al. The clinical outcome of pazopanib treatment in Japanese patients with relapsed soft tissue sarcoma: a Japanese Musculoskeletal Oncology Group (JMOG) study. Cancer. 2016; 122:1408–16.
Article15. Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010; 29:625–34.
Article16. Zelzer E, Levy Y, Kahana C, Shilo BZ, Rubinstein M, Cohen B. Insulin induces transcription of target genes through the hypoxia-inducible factor HIF-1alpha/ARNT. EMBO J. 1998; 17:5085–94.17. AACR Project GENIE Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017; 7:818–31.18. Koehler K, Liebner D, Chen JL. TP53 mutational status is predictive of pazopanib response in advanced sarcomas. Ann Oncol. 2016; 27:539–43.
Article19. Wheler JJ, Janku F, Naing A, Li Y, Stephen B, Zinner R, et al. TP53 alterations correlate with response to VEGF/VEGFR inhibitors: implications for targeted therapeutics. Mol Cancer Ther. 2016; 15:2475–85.20. Petitprez F, de Reynies A, Keung EZ, Chen TW, Sun CM, Calderaro J, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020; 577:556–60.
Article21. Allen E, Jabouille A, Rivera LB, Lodewijckx I, Missiaen R, Steri V, et al. Combined antiangiogenic and anti-PD-L1 therapy stimulates tumor immunity through HEV formation. Sci Transl Med. 2017; 9:eaak9679.
Article22. Aggen DH, Drake CG, Rini BI. Targeting PD-1 or PD-L1 in metastatic kidney cancer: combination therapy in the first-line setting. Clin Cancer Res. 2020; 26:2087–95.
Article23. Wilky BA, Trucco MM, Subhawong TK, Florou V, Park W, Kwon D, et al. Axitinib plus pembrolizumab in patients with advanced sarcomas including alveolar soft-part sarcoma: a single-centre, single-arm, phase 2 trial. Lancet Oncol. 2019; 20:837–48.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Durable Response to Pazopanib in a Patient with Metastatic Alveolar Soft Part Sarcoma
- Comparative Evaluation of Second-Line Chemotherapy Agents for Advanced Soft Tissue Sarcoma: Gemcitabine/Docetaxel, Pazopanib, and Alternatives
- Treatment of a Patient with Kaposi's Sarcoma Arising during Hemodialysis with the Multikinase Inhibitor Pazopanib
- Pazopanib monotherapy in the treatment of pretreated, metastatic uterine sarcoma: a single-center retrospective study
- Trial of Pazopanib in Proximal-type Epithelioid Sarcoma