J Korean Med Sci.
2023 Mar;38(10):e73. 10.3346/jkms.2023.38.e73.
Changing Epidemiology of Pathogenic Bacteria Over the Past 20 Years in Korea
- Affiliations
-
- 1Department of Laboratory Medicine, Hanyang University Guri Hospital, Hanyang University College of Medicine, Guri, Korea
- 2Department of Laboratory Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 3Department of Laboratory Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 4Department of Laboratory Medicine, Gyeongsang National University College of Medicine, Jinju, Korea
- 5Department of Laboratory Medicine and Research Institute of Bacterial Resistance, Yonsei University Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 6Department of Laboratory Medicine, Dongguk University Gyeongju Hospital, Dongguk University College of Medicine, Gyeongju, Korea
- 7Department of Laboratory Medicine, Keimyung University Dongsan Hospital, Keimyung University School of Medicine, Daegu, Korea
- 8Department of Laboratory Medicine, Yonsei University Wonju Severance Christian Hospital, Yonsei University Wonju College of Medicine, Wonju, Korea
- 9Department of Laboratory Medicine, Chonnam National University Hospital, Chonnam National University Medical School, Gwangju, Korea
- 10Department of Laboratory Medicine, Chonbuk National University Hospital, Chonbuk National University Medical School, Jeonju, Korea
- 11Department of Laboratory Medicine, Eulji University Hospital, Eulji University School of Medicine, Daejeon, Korea
- 12Department of Laboratory Medicine, Seoul Metropolitan Government-Seoul National University Boramae Medical Center, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2540431
- DOI: http://doi.org/10.3346/jkms.2023.38.e73
Abstract
- Background
The epidemiology of pathogenic bacteria varies according to the socioeconomic status and antimicrobial resistance status. However, longitudinal epidemiological studies to evaluate the changes in species distribution and antimicrobial susceptibility of pathogenic bacteria nationwide are lacking. We retrospectively investigated the nationwide trends in species distribution and antimicrobial susceptibility of pathogenic bacteria over the last 20 years in Korea.
Methods
From 1997 to 2016, annual cumulative antimicrobial susceptibility and species distribution data were collected from 12 university hospitals in five provinces and four metropolitan cities in South Korea.
Results
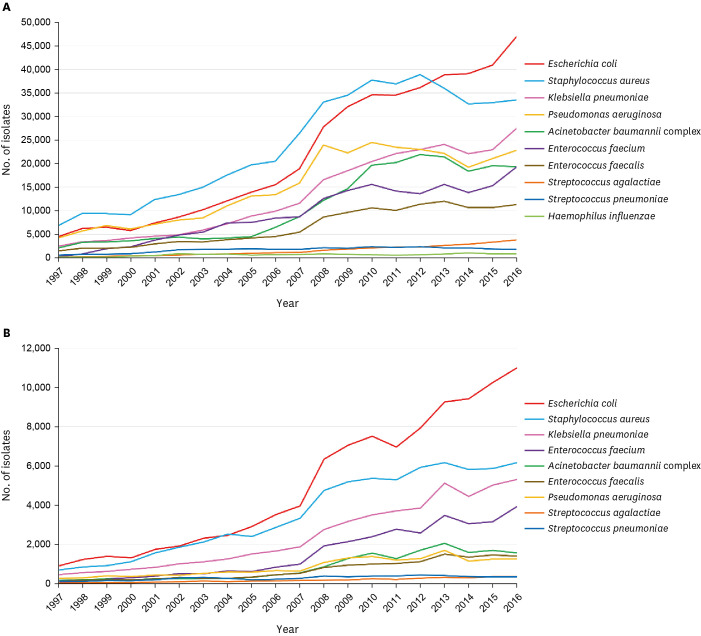
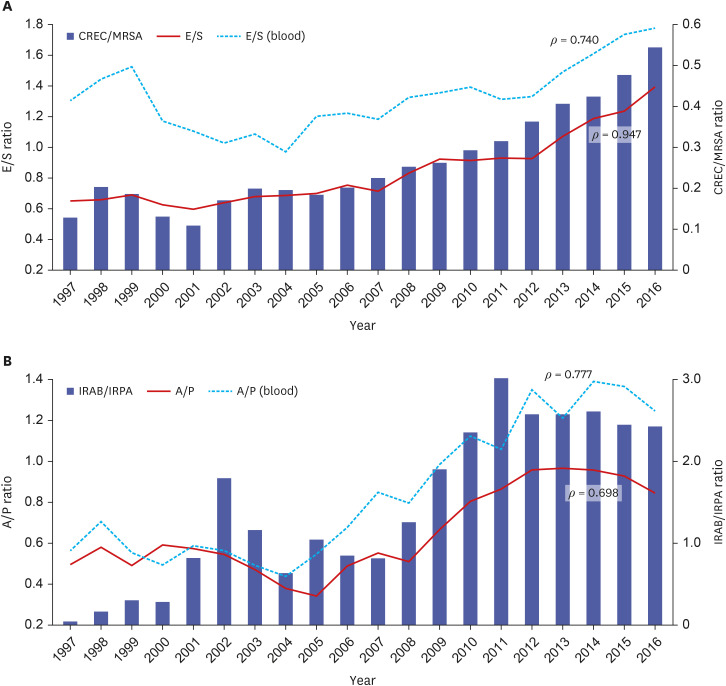
The prevalence of Staphylococcus aureus was the highest (13.1%) until 2012 but decreased to 10.3% in 2016, consistent with the decrease in oxacillin resistance from 76.1% in 2008 to 62.5% in 2016. While the cefotaxime resistance of Escherichia coli increased from 9.0% in 1997 to 34.2% in 2016, E. coli became the most common species since 2013, accounting for 14.5% of all isolates in 2016. Pseudomonas aeruginosa and Acinetobacter baumannii rose to third and fifth places in 2008 and 2010, respectively, while imipenem resistance increased from 13.9% to 30.8% and 0.7% to 73.5% during the study period, respectively. Streptococcus agalactiae became the most common pathogenic streptococcal species in 2016, as the prevalence of Streptococcus pneumoniae decreased since 2010. During the same period, pneumococcal penicillin susceptibility decreased to 79.0%, and levofloxacin susceptibility of S. agalactiae decreased to 77.1% in 2016.
Conclusion
The epidemiology of pathogenic bacteria has changed significantly over the past 20 years according to trends in antimicrobial resistance in Korea. Efforts to confine antimicrobial resistance would change the epidemiology of pathogenic bacteria and, consequently, the diagnosis and treatment of infectious diseases.
Figure
Cited by 1 articles
-
Changing Genotypic Distribution, Antimicrobial Susceptibilities, and Risk Factors of Urinary Tract Infection Caused by Carbapenemase-Producing
Pseudomonas aeruginosa
Seri Jeong, Kibum Jeon, Nuri Lee, Min-Jeong Park, Wonkeun Song
Ann Lab Med. 2024;44(1):38-46. doi: 10.3343/alm.2024.44.1.38.
Reference
-
1. Murray CJ, Lopez AD. Global mortality, disability, and the contribution of risk factors: Global Burden of Disease Study. Lancet. 1997; 349(9063):1436–1442. PMID: 9164317.2. Cohen ML. Changing patterns of infectious disease. Nature. 2000; 406(6797):762–767. PMID: 10963605.3. Chambers HF. The changing epidemiology of Staphylococcus aureus? Emerg Infect Dis. 2001; 7(2):178–182. PMID: 11294701.4. Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012; 18(5):263–272. PMID: 22480775.5. World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report Early Implementation 2016–2017. Updated 2018. Accessed October 15, 2020. https://www.who.int/publications/i/item/9789241513449 .6. Choe YJ, Choe SA, Cho SI. Trends in infectious disease mortality, South Korea, 1983-2015. Emerg Infect Dis. 2018; 24(2):320–327. PMID: 29350153.7. O’Brien TF, Stelling JM. WHONET: removing obstacles to the full use of information about antimicrobial resistance. Diagn Microbiol Infect Dis. 1996; 25(4):162–168. PMID: 8937840.8. Ramanan P, Bryson AL, Binnicker MJ, Pritt BS, Patel R. Syndromic panel-based testing in clinical microbiology. Clin Microbiol Rev. 2018; 31(1):e00024-17. PMID: 29142077.9. Diekema DJ, Hsueh PR, Mendes RE, Pfaller MA, Rolston KV, Sader HS, et al. The microbiology of bloodstream infection: 20-year trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother. 2019; 63(7):e00355-19. PMID: 31010862.10. Choi JY, Kwak YG, Yoo H, Lee SO, Kim HB, Han SH, et al. Trends in the distribution and antimicrobial susceptibility of causative pathogens of device-associated infection in Korean intensive care units from 2006 to 2013: results from the Korean Nosocomial Infections Surveillance System (KONIS). J Hosp Infect. 2016; 92(4):363–371. PMID: 26876746.11. Lee K, Kim MN, Kim JS, Hong HL, Kang JO, Shin JH, et al. Further increases in carbapenem-, amikacin-, and fluoroquinolone-resistant isolates of Acinetobacter spp. and P. aeruginosa in Korea: KONSAR study 2009. Yonsei Med J. 2011; 52(5):793–802. PMID: 21786445.12. Yong D, Shin HB, Kim YK, Cho J, Lee WG, Ha GY, et al. Increase in the prevalence of carbapenem-resistant Acinetobacter isolates and ampicillin-resistant non-typhoidal salmonella species in Korea: a KONSAR study conducted in 2011. Infect Chemother. 2014; 46(2):84–93. PMID: 25024870.13. Kim D, Ahn JY, Lee CH, Jang SJ, Lee H, Yong D, et al. Increasing resistance to extended-spectrum cephalosporins, fluoroquinolone, and carbapenem in gram-negative bacilli and the emergence of carbapenem non-susceptibility in Klebsiella pneumoniae: analysis of Korean Antimicrobial Resistance Monitoring System (KARMS) data from 2013 to 2015. Ann Lab Med. 2017; 37(3):231–239. PMID: 28224769.14. Kim H, Kim ES, Lee SC, Yang E, Kim HS, Sung H, et al. Decreased incidence of methicillin-resistant Staphylococcus aureus bacteremia in intensive care units: a 10-year clinical, microbiological, and genotypic analysis in a tertiary hospital. Antimicrob Agents Chemother. 2020; 64(10):e01082-20. PMID: 32747360.15. Song W, Lee H, Lee K, Jeong SH, Bae IK, Kim JS, et al. CTX-M-14 and CTX-M-15 enzymes are the dominant type of extended-spectrum beta-lactamase in clinical isolates of Escherichia coli from Korea. J Med Microbiol. 2009; 58(Pt 2):261–266. PMID: 19141747.16. Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011; 66(1):1–14. PMID: 21081548.17. Lee MY, Choi HJ, Choi JY, Song M, Song Y, Kim SW, et al. Dissemination of ST131 and ST393 community-onset, ciprofloxacin-resistant Escherichia coli clones causing urinary tract infections in Korea. J Infect. 2010; 60(2):146–153. PMID: 19932131.18. Park SH, Byun JH, Choi SM, Lee DG, Kim SH, Kwon JC, et al. Molecular epidemiology of extended-spectrum β-lactamase-producing Escherichia coli in the community and hospital in Korea: emergence of ST131 producing CTX-M-15. BMC Infect Dis. 2012; 12(1):149. PMID: 22747570.19. Moellering RC Jr. NDM-1--a cause for worldwide concern. N Engl J Med. 2010; 363(25):2377–2379. PMID: 21158655.20. Yoon EJ, Yang JW, Kim JO, Lee H, Lee KJ, Jeong SH. Carbapenemase-producing Enterobacteriaceae in South Korea: a report from the National Laboratory Surveillance System. Future Microbiol. 2018; 13(7):771–783. PMID: 29478336.21. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017; 215(suppl_1):S28–S36. PMID: 28375512.22. Sader HS, Castanheira M, Arends SJ, Goossens H, Flamm RK. Geographical and temporal variation in the frequency and antimicrobial susceptibility of bacteria isolated from patients hospitalized with bacterial pneumonia: results from 20 years of the SENTRY Antimicrobial Surveillance Program (1997-2016). J Antimicrob Chemother. 2019; 74(6):1595–1606. PMID: 30843070.23. Shortridge D, Gales AC, Streit JM, Huband MD, Tsakris A, Jones RN. Geographic and temporal patterns of antimicrobial resistance in Pseudomonas aeruginosa over 20 years from the SENTRY Antimicrobial Surveillance Program, 1997-2016. Open Forum Infect Dis. 2019; 6(Suppl 1):S63–S68. PMID: 30895216.24. Lee Y, Kim YR, Kim J, Park YJ, Song W, Shin JH, et al. Increasing prevalence of blaOXA-23-carrying Acinetobacter baumannii and the emergence of blaOXA-182-carrying Acinetobacter nosocomialis in Korea. Diagn Microbiol Infect Dis. 2013; 77(2):160–163. PMID: 23891219.25. Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011; 35(5):736–755. PMID: 21303394.26. Bae IK, Suh B, Jeong SH, Wang KK, Kim YR, Yong D, et al. Molecular epidemiology of Pseudomonas aeruginosa clinical isolates from Korea producing β-lactamases with extended-spectrum activity. Diagn Microbiol Infect Dis. 2014; 79(3):373–377. PMID: 24792837.27. Hong JS, Yoon EJ, Lee H, Jeong SH, Lee K. Clonal dissemination of Pseudomonas aeruginosa sequence type 235 isolates carrying blaIMP-6 and emergence of blaGES-24 and blaIMP-10 on novel genomic islands PAGI-15 and -16 in South Korea. Antimicrob Agents Chemother. 2016; 60(12):7216–7223. PMID: 27671068.28. Mendes RE, Castanheira M, Farrell DJ, Flamm RK, Sader HS, Jones RN. Longitudinal (2001-14) analysis of enterococci and VRE causing invasive infections in European and US hospitals, including a contemporary (2010-13) analysis of oritavancin in vitro potency. J Antimicrob Chemother. 2016; 71(12):3453–3458. PMID: 27609052.29. Gao W, Howden BP, Stinear TP. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr Opin Microbiol. 2018; 41:76–82. PMID: 29227922.30. Richter SS, Diekema DJ, Heilmann KP, Dohrn CL, Riahi F, Doern GV. Changes in pneumococcal serotypes and antimicrobial resistance after introduction of the 13-valent conjugate vaccine in the United States. Antimicrob Agents Chemother. 2014; 58(11):6484–6489. PMID: 25136018.31. Park DC, Kim SH, Yong D, Suh IB, Kim YR, Yi J, et al. Serotype distribution and antimicrobial resistance of invasive and noninvasive Streptococcus pneumoniae Isolates in Korea between 2014 and 2016. Ann Lab Med. 2019; 39(6):537–544. PMID: 31240881.32. Kim SH, Song JH, Chung DR, Thamlikitkul V, Yang Y, Wang H, et al. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Antimicrob Agents Chemother. 2012; 56(3):1418–1426. PMID: 22232285.33. Rhie K, Choi EH, Cho EY, Lee J, Kang JH, Kim DS, et al. Etiology of invasive bacterial infections in immunocompetent children in Korea (2006-2010): a retrospective multicenter study. J Korean Med Sci. 2018; 33(6):e45. PMID: 29349940.34. Matsubara K, Yamamoto G. Invasive group B streptococcal infections in a tertiary care hospital between 1998 and 2007 in Japan. Int J Infect Dis. 2009; 13(6):679–684. PMID: 19131262.35. Björnsdóttir ES, Martins ER, Erlendsdóttir H, Haraldsson G, Melo-Cristino J, Kristinsson KG, et al. Changing epidemiology of group B streptococcal infections among adults in Iceland: 1975-2014. Clin Microbiol Infect. 2016; 22(4):379.e9–e16.36. Lee K, Kim WJ, Kim DL, Ko HM, Baik SH, Kim MN, et al. Clinical characteristics of Streptococcus agalactiae bacteremia in adults living in Jeju Island. Ann Clin Microbiol. 2014; 17(1):9–13.37. Ki M, Srinivasan U, Oh KY, Kim MY, Shin JH, Hong HL, et al. Emerging fluoroquinolone resistance in Streptococcus agalactiae in South Korea. Eur J Clin Microbiol Infect Dis. 2012; 31(11):3199–3205. PMID: 22752224.38. Song JH, Jung KS, Kang MW, Kim DJ, Pai H, Suh GY, et al. Treatment guidelines for community-acquired pneumonia in Korea: an evidence-based approach to appropriate antimicrobial therapy. Tuberc Respir Dis. 2009; 67(4):281–302.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Enteric Bacteria Isolated from Diarrheal Patients in Korea in 2014
- Multidrug-Resistant Gram-Positive Bacterial Infections
- The Prevalence and Characteristics of Bacteria Causing Acute Diarrhea in Korea, 2012
- Reduced alveolar bone loss in rats immunized with Porphyromonas gingivalis heat shock protein
- Bacterial Contamination Conditions in Ambulances and their Equipment in South Korea