Cancer Res Treat.
2021 Oct;53(4):1042-1056. 10.4143/crt.2020.1208.
Inhibition of lncRNA KCNQ1OT1 Improves Apoptosis and Chemotherapy Drug Response in Small Cell Lung Cancer by TGF-β1 Mediated Epithelial-to-Mesenchymal Transition
- Affiliations
-
- 1Department of Pathology, Zhujiang Hospital, Southern Medical University, Guangzhou, China
- 2Department of Medical Oncology,Provincial Clinical College, Fujian Medical University, Fujian provincial hospital, Fuzhou, China
- 3Department of Radiation Oncology, The First Affiliated Hospital of University of South China, Hengyang, China
- 4Department of Pathology, The Union Hospital of Fujian Medical University, Fuzhou, China
- 5Department of Pathology, Provincial Clinical College, Fujian Medical University, Fujian provincial hospital, Fuzhou, China
- 6Department of Cardiothoracic Surgery, Zhujiang Hospital, Southern Medical University, Guangzhou, China
- 7Guangdong Provincial Corps Hospital of Chinese People’s Armed Police Forces, Guangzhou Medical University, Guangzhou, China
- KMID: 2521580
- DOI: http://doi.org/10.4143/crt.2020.1208
Abstract
- Purpose
Drug resistance is one of the main causes of chemotherapy failure in patients with small cell lung cancer (SCLC), and extensive biological studies into chemotherapy drug resistance are required.
Materials and Methods
In this study, we performed lncRNA microarray, in vitro functional assays, in vivo models and cDNA microarray to evaluate the impact of lncRNA in SCLC chemoresistance.
Results
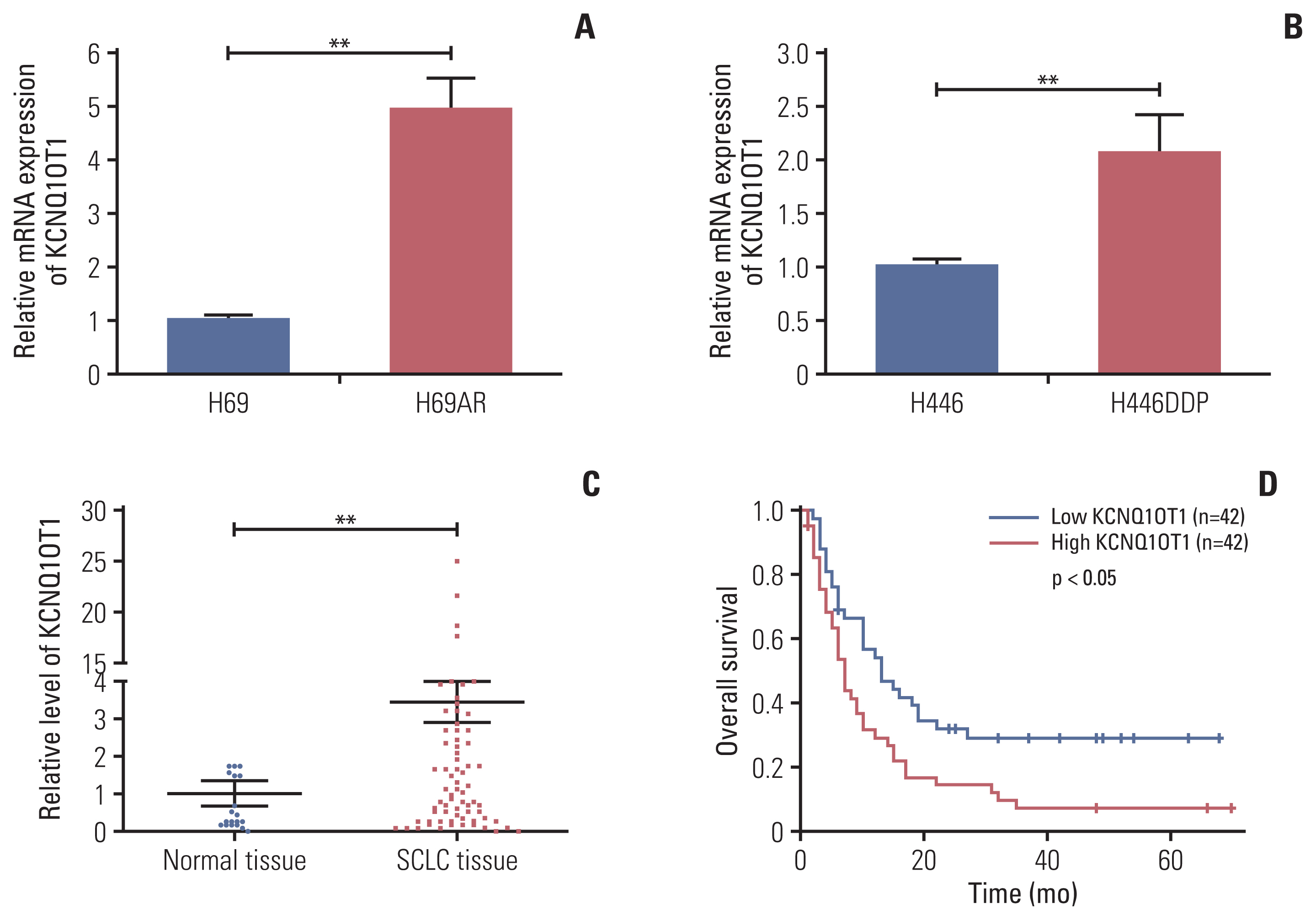
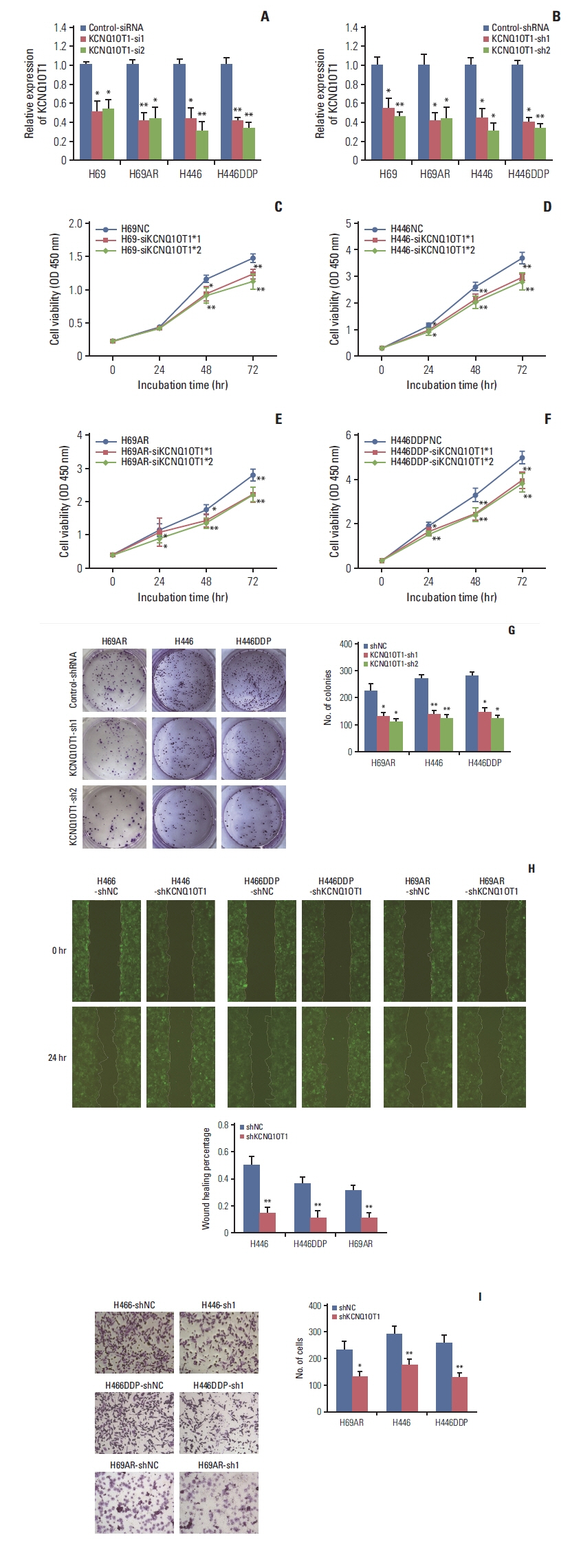
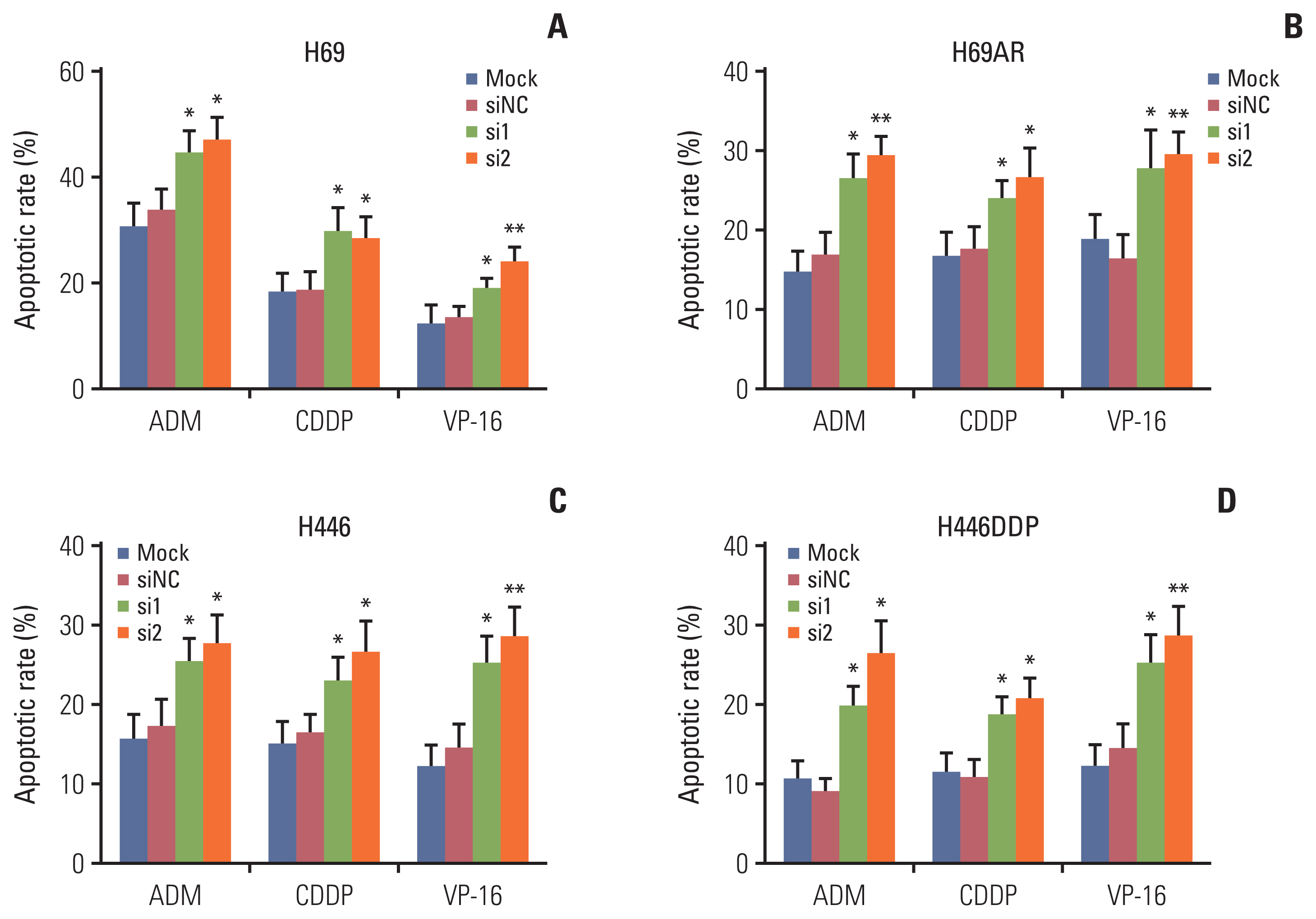
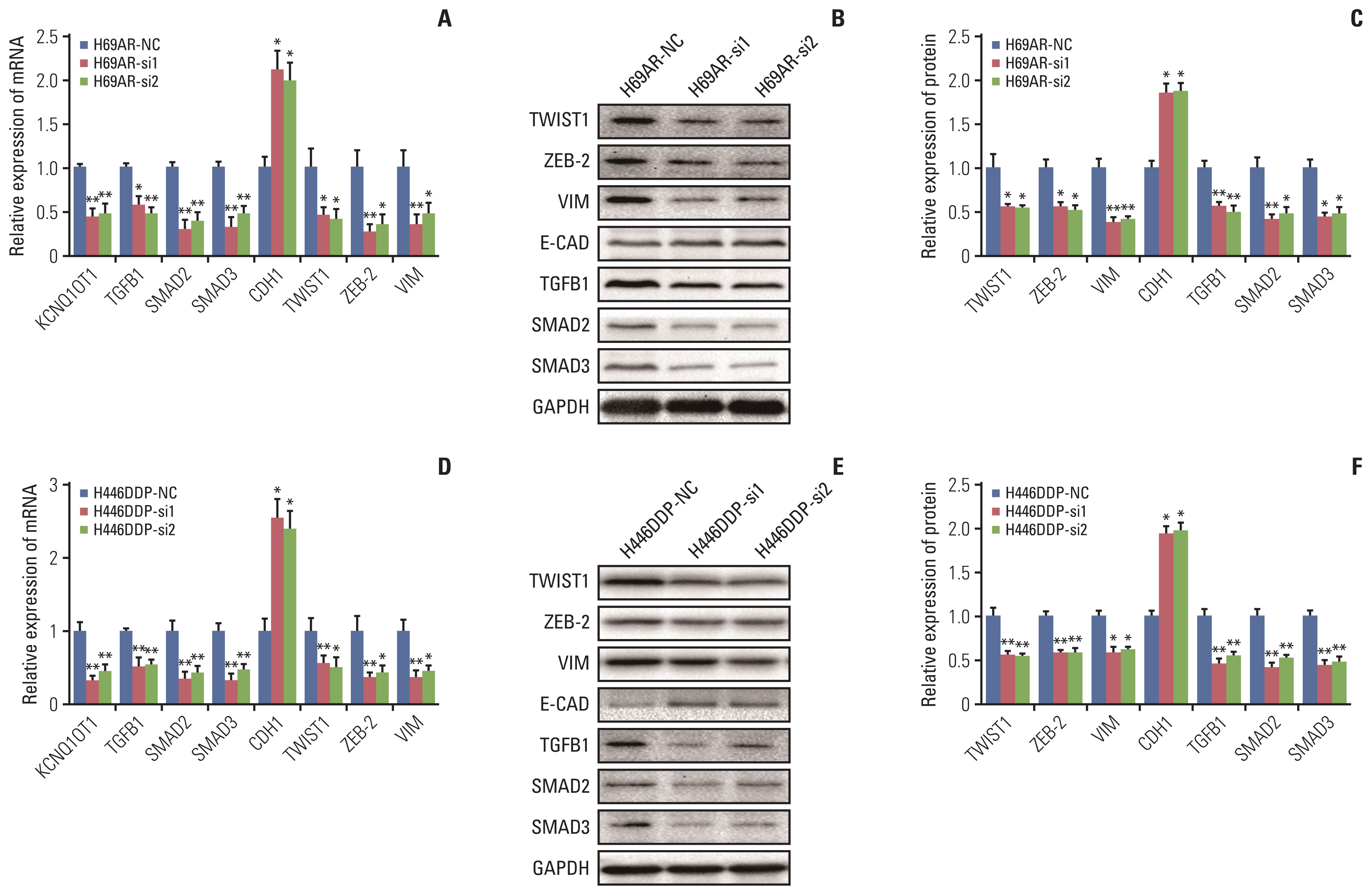
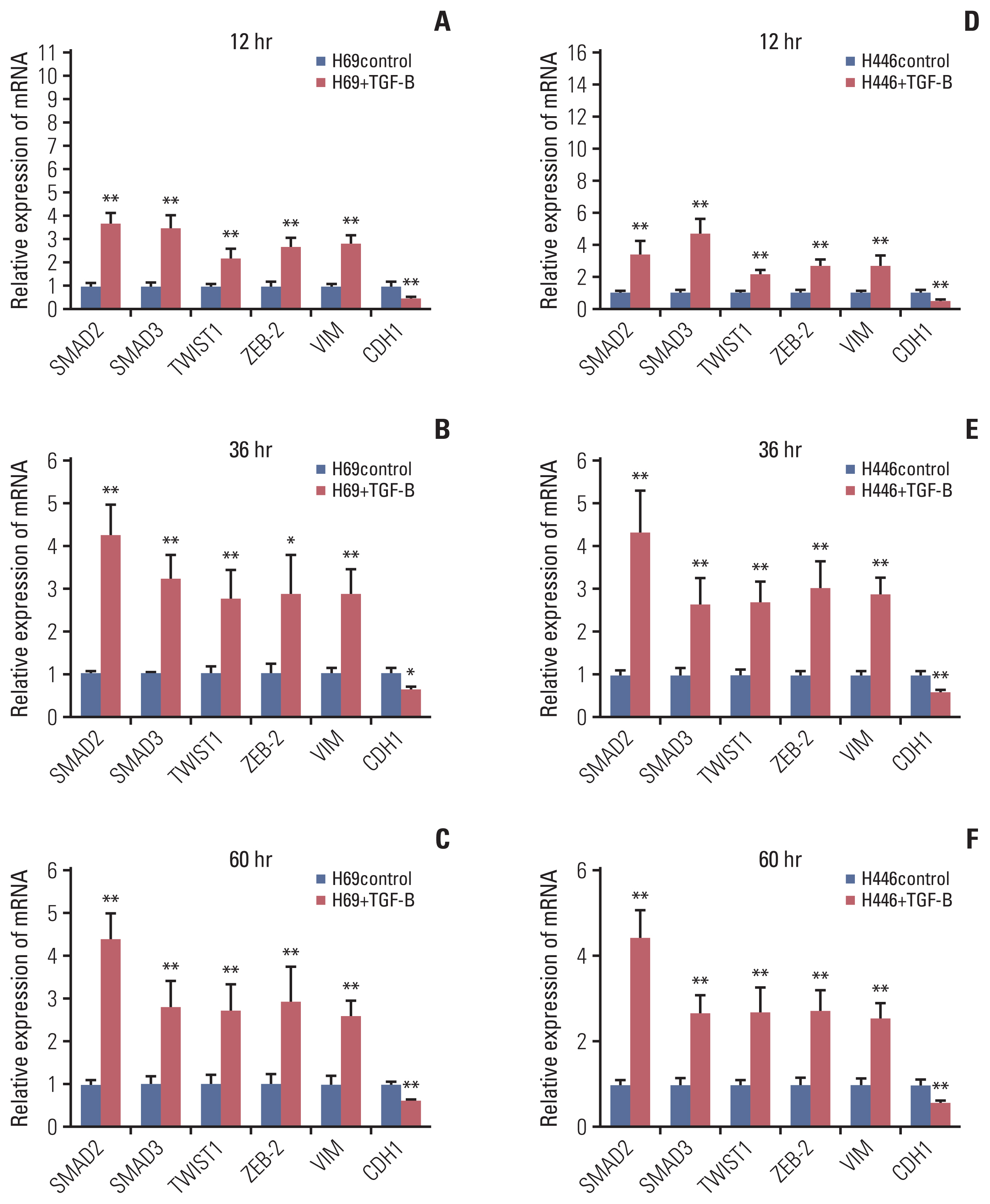
The results showed that KCNQ1OT1 expression was upregulated in SCLC tissues and was a poor prognostic factor for patients with SCLC. Knockdown of KCNQ1OT1 inhibited cell proliferation, migration, chemoresistance and promoted apoptosis of SCLC cells. Mechanistic investigation showed that KCNQ1OT1 can activate transforming growth factor-β1 mediated epithelial-to-mesenchymal transition in SCLC cells.
Conclusion
Taken together, our study revealed the role of KCNQ1OT1 in the progression and chemoresistance of SCLC, and suggested KCNQ1OT1 as a potential diagnostic and prognostic biomarker in SCLC clinical management.
Keyword
Figure
Reference
-
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015; 136:E359–86.
Article2. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012; 380:2095–128.3. Sabari JK, Lok BH, Laird JH, Poirier JT, Rudin CM. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol. 2017; 14:549–61.
Article4. Faivre-Finn C, Snee M, Ashcroft L, Appel W, Barlesi F, Bhatnagar A, et al. Concurrent once-daily versus twice-daily chemoradiotherapy in patients with limited-stage small-cell lung cancer (CONVERT): an open-label, phase 3, randomised, superiority trial. Lancet Oncol. 2017; 18:1116–25.
Article5. Gaspar LE, McNamara EJ, Gay EG, Putnam JB, Crawford J, Herbst RS, et al. Small-cell lung cancer: prognostic factors and changing treatment over 15 years. Clin Lung Cancer. 2012; 13:115–22.
Article6. Wolin SL, Maquat LE. Cellular RNA surveillance in health and disease. Science. 2019; 366:822–7.
Article7. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019; 179:1033–55.
Article8. Li Z, Zhang J, Liu X, Li S, Wang Q, Di C, et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun. 2018; 9:1572.
Article9. Zhuo W, Liu Y, Li S, Guo D, Sun Q, Jin J, et al. Long noncoding RNA GMAN, up-regulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of ephrin A1 by competitively binding GMAN-AS. Gastroenterology. 2019; 156:676–91.
Article10. Mendell JT. Targeting a long noncoding RNA in breast cancer. N Engl J Med. 2016; 374:2287–9.
Article11. Fang S, Gao H, Tong Y, Yang J, Tang R, Niu Y, et al. Long noncoding RNA-HOTAIR affects chemoresistance by regulating HOXA1 methylation in small cell lung cancer cells. Lab Invest. 2016; 96:60–8.
Article12. Sun Y, Hu B, Wang Q, Ye M, Qiu Q, Zhou Y, et al. Long non-coding RNA HOTTIP promotes BCL-2 expression and induces chemoresistance in small cell lung cancer by sponging miR-216a. Cell Death Dis. 2018; 9:85.
Article13. Sun Y, Zhou Y, Bai Y, Wang Q, Bao J, Luo Y, et al. A long non-coding RNA HOTTIP expression is associated with disease progression and predicts outcome in small cell lung cancer patients. Mol Cancer. 2017; 16:162.
Article14. Zeng F, Wang Q, Wang S, Liang S, Huang W, Guo Y, et al. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate Etk expression. Oncogene. 2020; 39:293–307.
Article15. Luo ZP, Jin H. Effects of LncRNA KCNQ1OT1 on proliferation and migration of ovarian cancer cells by Wnt/beta-catenin. Eur Rev Med Pharmacol Sci. 2019; 23:8788–94.16. Guo B, Zhang Q, Wang H, Chang P, Tao K. KCNQ1OT1 promotes melanoma growth and metastasis. Aging (Albany NY). 2018; 10:632–44.
Article17. Xian D, Zhao Y. LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR-760/PPP1R1B via the cAMP signalling pathway. J Cell Mol Med. 2019; 23:3808–23.18. Wang J, Zhang H, Situ J, Li M, Sun H. KCNQ1OT1 aggravates cell proliferation and migration in bladder cancer through modulating miR-145-5p/PCBP2 axis. Cancer Cell Int. 2019; 19:325.
Article19. Zheng L, Zhang FX, Wang LL, Hu HL, Lian YD. LncRNA KCNQ1OT1 is overexpressed in non-small cell lung cancer and its expression level is related to clinicopathology. Eur Rev Med Pharmacol Sci. 2019; 23:6944–50.20. Sun X, Xin Y, Wang M, Li S, Miao S, Xuan Y, et al. Overexpression of long non-coding RNA KCNQ1OT1 is related to good prognosis via inhibiting cell proliferation in non-small cell lung cancer. Thorac Cancer. 2018; 9:523–31.
Article21. Li Y, Li C, Li D, Yang L, Jin J, Zhang B. lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway. Onco Targets Ther. 2019; 12:2649–60.22. Hu H, Yang L, Li L, Zeng C. Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ABCC1 axis. Biochem Biophys Res Commun. 2018; 503:2400–6.23. Fujita K, Iwama H, Oura K, Tadokoro T, Samukawa E, Sakamoto T, et al. Cancer therapy due to apoptosis: galectin-9. Int J Mol Sci. 2017; 18:74.
Article24. Park JH, Kim YH, Park EH, Lee SJ, Kim H, Kim A, et al. Effects of metformin and phenformin on apoptosis and epithelial-mesenchymal transition in chemoresistant rectal cancer. Cancer Sci. 2019; 110:2834–45.
Article25. Rouhrazi H, Turgan N, Oktem G. Zoledronic acid overcomes chemoresistance by sensitizing cancer stem cells to apoptosis. Biotech Histochem. 2018; 93:77–88.
Article26. Sun H, Li Y, Kong H, Dai S, Qian H. Dysregulation of KCNQ1OT1 promotes cholangiocarcinoma progression via miR-140-5p/SOX4 axis. Arch Biochem Biophys. 2018; 658:7–15.
Article27. Liu H, Chen R, Kang F, Lai H, Wang Y. KCNQ1OT1 promotes ovarian cancer progression via modulating MIR-142-5p/CAPN10 axis. Mol Genet Genomic Med. 2020; 8:e1077.
Article28. Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules. 2016; 21:965.
Article29. Zhu L, Fu X, Chen X, Han X, Dong P. M2 macrophages induce EMT through the TGF-beta/Smad2 signaling pathway. Cell Biol Int. 2017; 41:960–8.30. Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009; 19:156–72.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Ginsenoside Rb1 Attenuates TGF-β1-Induced MUC4/5AC Expression and Epithelial-Mesenchymal Transition in Human Airway Epithelial Cells
- Epithelial Mesenchymal Transition in Drug Resistance and Metastasis of Lung Cancer
- Apoptosis and Cell Cycle Arrest with EGF, TGF- a and TGF- 8 in Cervical Cancer Cell Lines
- Trefoil Factor 1 Suppresses Epithelial-mesenchymal Transition through Inhibition of TGF-beta Signaling in Gastric Cancer Cells
- Parthenolide inhibits transforming growth factor β1-induced epithelial-mesenchymal transition in colorectal cancer cells