Allergy Asthma Immunol Res.
2020 May;12(3):399-411. 10.4168/aair.2020.12.3.399.
Understanding the Molecular Mechanisms of Asthma through Transcriptomics
- Affiliations
-
- 1The Channing Division of Network Medicine, Department of Medicine, Brigham & Women's Hospital and Harvard Medical School, Boston, MA, USA. restw@channing.harvard.edu
- 2Department of Internal Medicine, Seoul National University College of Medicine, Seoul, Korea.
- 3Partners Center for Personalized Medicine, Partners Health Care, Boston, MA, USA.
- KMID: 2471225
- DOI: http://doi.org/10.4168/aair.2020.12.3.399
Abstract
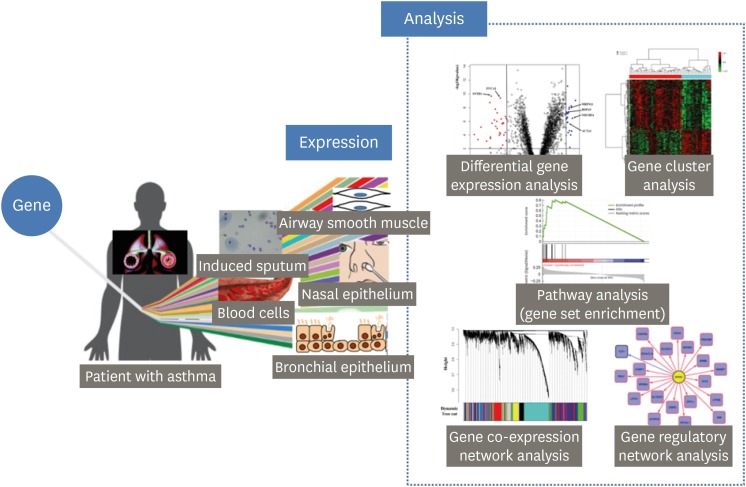
- The transcriptome represents the complete set of RNA transcripts that are produced by the genome under a specific circumstance or in a specific cell. High-throughput methods, including microarray and bulk RNA sequencing, as well as recent advances in biostatistics based on machine learning approaches provides a quick and effective way of identifying novel genes and pathways related to asthma, which is a heterogeneous disease with diverse pathophysiological mechanisms. In this manuscript, we briefly review how to analyze transcriptome data and then provide a summary of recent transcriptome studies focusing on asthma pathogenesis and asthma drug responses. Studies reviewed here are classified into 2 classes based on the tissues utilized: blood and airway cells.
Keyword
MeSH Terms
Figure
Reference
-
1. Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007; 448:470–473. PMID: 17611496.2. Diao G, Vidyashankar AN. Assessing genome-wide statistical significance for large p small n problems. Genetics. 2013; 194:781–783. PMID: 23666935.3. Lee JH, Kaminski N, Dolganov G, Grunig G, Koth L, Solomon C, et al. Interleukin-13 induces dramatically different transcriptional programs in three human airway cell types. Am J Respir Cell Mol Biol. 2001; 25:474–485. PMID: 11694453.
Article4. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102:15545–15550. PMID: 16199517.
Article5. Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, et al. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004; 101:6570–6575. PMID: 15100410.
Article6. Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature. 2007; 450:736–740. PMID: 18046414.
Article7. Tamayo P, Steinhardt G, Liberzon A, Mesirov JP. The limitations of simple gene set enrichment analysis assuming gene independence. Stat Methods Med Res. 2016; 25:472–487. PMID: 23070592.
Article8. Sonawane AR, Weiss ST, Glass K, Sharma A. Network medicine in the age of biomedical big data. Front Genet. 2019; 10:294. PMID: 31031797.
Article9. Serin EA, Nijveen H, Hilhorst HW, Ligterink W. Learning from co-expression networks: possibilities and challenges. Front Plant Sci. 2016; 7:444. PMID: 27092161.
Article10. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008; 9:559. PMID: 19114008.
Article11. Reverter A, Chan EK. Combining partial correlation and an information theory approach to the reversed engineering of gene co-expression networks. Bioinformatics. 2008; 24:2491–2497. PMID: 18784117.
Article12. Roy S, Bhattacharyya DK, Kalita JK. Reconstruction of gene co-expression network from microarray data using local expression patterns. BMC Bioinformatics. 2014; 15 Suppl 7:S10.
Article13. van Dam S, Võsa U, van der Graaf A, Franke L, de Magalhães JP. Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform. 2018; 19:575–592. PMID: 28077403.
Article14. Kelly RS, Chawes BL, Blighe K, Virkud YV, Croteau-Chonka DC, McGeachie MJ, et al. An integrative transcriptomic and metabolomic study of lung function in children with asthma. Chest. 2018; 154:335–348. PMID: 29908154.
Article15. Aldrich J. Correlations genuine and spurious in Pearson and Yule. Stat Sci. 1995; 10:364–376.
Article16. Marbach D, Lamparter D, Quon G, Kellis M, Kutalik Z, Bergmann S. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat Methods. 2016; 13:366–370. PMID: 26950747.
Article17. Caramori G, Casolari P, Adcock I. Role of transcription factors in the pathogenesis of asthma and COPD. Cell Commun Adhes. 2013; 20:21–40. PMID: 23472830.
Article18. Glass K, Huttenhower C, Quackenbush J, Yuan GC. Passing messages between biological networks to refine predicted interactions. PLoS One. 2013; 8:e64832. PMID: 23741402.
Article19. Schlauch D, Paulson JN, Young A, Glass K, Quackenbush J. Estimating gene regulatory networks with pandaR. Bioinformatics. 2017; 33:2232–2234. PMID: 28334344.
Article20. Qiu W, Guo F, Glass K, Yuan GC, Quackenbush J, Zhou X, et al. Differential connectivity of gene regulatory networks distinguishes corticosteroid response in asthma. J Allergy Clin Immunol. 2018; 141:1250–1258. PMID: 28736268.
Article21. Bar-Joseph Z, Gitter A, Simon I. Studying and modelling dynamic biological processes using time-series gene expression data. Nat Rev Genet. 2012; 13:552–564. PMID: 22805708.
Article22. Oh S, Song S, Dasgupta N, Grabowski G. The analytical landscape of static and temporal dynamics in transcriptome data. Front Genet. 2014; 5:35. PMID: 24600473.
Article23. Más P. Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol. 2008; 18:273–281. PMID: 18468438.24. Aryee MJ, Gutiérrez-Pabello JA, Kramnik I, Maiti T, Quackenbush J. An improved empirical bayes approach to estimating differential gene expression in microarray time-course data: BETR (Bayesian Estimation of Temporal Regulation). BMC Bioinformatics. 2009; 10:409. PMID: 20003283.
Article25. Nueda MJ, Tarazona S, Conesa A. Next maSigPro: updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics. 2014; 30:2598–2602. PMID: 24894503.
Article26. Leng N, Li Y, McIntosh BE, Nguyen BK, Duffin B, Tian S, et al. EBSeq-HMM: a Bayesian approach for identifying gene-expression changes in ordered RNA-seq experiments. Bioinformatics. 2015; 31:2614–2622. PMID: 25847007.
Article27. Kontou PI, Pavlopoulou A, Bagos PG. Methods of analysis and meta-analysis for identifying differentially expressed genes. Methods Mol Biol. 2018; 1793:183–210. PMID: 29876898.
Article28. Moher D, Liberati A, Tetzlaff J, Altman DG. PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010; 8:336–341. PMID: 20171303.
Article29. Waldron L, Riester M. Meta-analysis in gene expression studies. Methods Mol Biol. 2016; 1418:161–176. PMID: 27008014.
Article30. Sweeney TE, Haynes WA, Vallania F, Ioannidis JP, Khatri P. Methods to increase reproducibility in differential gene expression via meta-analysis. Nucleic Acids Res. 2017; 45:e1. PMID: 27634930.
Article31. Rau A, Marot G, Jaffrézic F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinformatics. 2014; 15:91. PMID: 24678608.
Article32. Conlon EM, Song JJ, Liu JS. Bayesian models for pooling microarray studies with multiple sources of replications. BMC Bioinformatics. 2006; 7:247. PMID: 16677390.
Article33. Zhou G, Soufan O, Ewald J, Hancock RE, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019; 47:W234–W241. PMID: 30931480.
Article34. Sharov AA, Schlessinger D, Ko MS. ExAtlas: an interactive online tool for meta-analysis of gene expression data. J Bioinform Comput Biol. 2015; 13:1550019. PMID: 26223199.
Article35. Chelly J, Kaplan JC, Maire P, Gautron S, Kahn A. Transcription of the dystrophin gene in human muscle and non-muscle tissue. Nature. 1988; 333:858–860. PMID: 3290682.36. Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006; 147:126–132. PMID: 16503242.
Article37. Yeh YL, Su MW, Chiang BL, Yang YH, Tsai CH, Lee YL. Genetic profiles of transcriptomic clusters of childhood asthma determine specific severe subtype. Clin Exp Allergy. 2018; 48:1164–1172. PMID: 29758111.
Article38. Croteau-Chonka DC, Qiu W, Martinez FD, Strunk RC, Lemanske RF Jr, Liu AH, et al. Gene expression profiling in blood provides reproducible molecular insights into asthma control. Am J Respir Crit Care Med. 2017; 195:179–188. PMID: 27494826.
Article39. Bjornsdottir US, Holgate ST, Reddy PS, Hill AA, McKee CM, Csimma CI, et al. Pathways activated during human asthma exacerbation as revealed by gene expression patterns in blood. PLoS One. 2011; 6:e21902. PMID: 21779351.
Article40. Altman MC, Gill MA, Whalen E, Babineau DC, Shao B, Liu AH, et al. Transcriptome networks identify mechanisms of viral and nonviral asthma exacerbations in children. Nat Immunol. 2019; 20:637–651. PMID: 30962590.
Article41. Persson H, Kwon AT, Ramilowski JA, Silberberg G, Söderhäll C, Orsmark-Pietras C, et al. Transcriptome analysis of controlled and therapy-resistant childhood asthma reveals distinct gene expression profiles. J Allergy Clin Immunol. 2015; 136:638–648. PMID: 25863981.
Article42. Bigler J, Boedigheimer M, Schofield JP, Skipp PJ, Corfield J, Rowe A, et al. A severe asthma disease signature from gene expression profiling of peripheral blood from U-BIOPRED cohorts. Am J Respir Crit Care Med. 2017; 195:1311–1320. PMID: 27925796.
Article43. Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X, et al. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol. 2012; 129:95–103. PMID: 21917308.44. Sawada R, Iwata M, Tabei Y, Yamato H, Yamanishi Y. Predicting inhibitory and activatory drug targets by chemically and genetically perturbed transcriptome signatures. Sci Rep. 2018; 8:156. PMID: 29317676.
Article45. Qiu W, Rogers AJ, Damask A, Raby BA, Klanderman BJ, Duan QL, et al. Pharmacogenomics: novel loci identification via integrating gene differential analysis and eQTL analysis. Hum Mol Genet. 2014; 23:5017–5024. PMID: 24770851.
Article46. Park HW, Dahlin A, Qiu W, Tantisira KG. Gene expression changes in lymphoblastoid cell lines and primary B cells by dexamethasone. Pharmacogenet Genomics. 2019; 29:58–64. PMID: 30562215.
Article47. Tsai YH, Parker JS, Yang IV, Kelada SNP. Meta-analysis of airway epithelium gene expression in asthma. Eur Respir J. 2018; 51:1701962. PMID: 29650561.
Article48. Modena BD, Bleecker ER, Busse WW, Erzurum SC, Gaston BM, Jarjour NN, et al. Gene expression correlated with severe asthma characteristics reveals heterogeneous mechanisms of severe disease. Am J Respir Crit Care Med. 2017; 195:1449–1463. PMID: 27984699.
Article49. Weathington N, O'Brien ME, Radder J, Whisenant TC, Bleecker ER, Busse WW, et al. BAL cell gene expression in severe asthma reveals mechanisms of severe disease and influences of medications. Am J Respir Crit Care Med. 2019; 200:837–856. PMID: 31161938.
Article50. Nicholas B, Djukanović R. Induced sputum: a window to lung pathology. Biochem Soc Trans. 2009; 37:868–872. PMID: 19614609.
Article51. Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011; 127:153–160. PMID: 21211650.
Article52. Kim BK, Lee HS, Sohn KH, Lee SY, Cho SH, Park HW. Different biological pathways are up-regulated in the elderly with asthma: sputum transcriptomic analysis. Allergy Asthma Immunol Res. 2019; 11:104–115. PMID: 30479081.
Article53. Peters MC, Ringel L, Dyjack N, Herrin R, Woodruff PG, Rios C, et al. A transcriptomic method to determine airway immune dysfunction in T2-high and T2-low asthma. Am J Respir Crit Care Med. 2019; 199:465–477. PMID: 30371106.
Article54. Sridhar S, Schembri F, Zeskind J, Shah V, Gustafson AM, Steiling K, et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomics. 2008; 9:259. PMID: 18513428.
Article55. Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O'Connor BP, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol. 2014; 133:670–678.e12. PMID: 24495433.
Article56. Pandey G, Pandey OP, Rogers AJ, Ahsen ME, Hoffman GE, Raby BA, et al. A nasal brush-based classifier of asthma identified by machine learning analysis of nasal RNA sequence data. Sci Rep. 2018; 8:8826. PMID: 29891868.
Article57. Hekking PP, Loza MJ, Pavlidis S, de Meulder B, Lefaudeux D, Baribaud F, et al. Pathway discovery using transcriptomic profiles in adult-onset severe asthma. J Allergy Clin Immunol. 2018; 141:1280–1290. PMID: 28756296.58. Woodruff PG. Gene expression in asthmatic airway smooth muscle. Proc Am Thorac Soc. 2008; 5:113–118. PMID: 18094093.
Article59. Yick CY, Zwinderman AH, Kunst PW, Grünberg K, Mauad T, Chowdhury S, et al. Gene expression profiling of laser microdissected airway smooth muscle tissue in asthma and atopy. Allergy. 2014; 69:1233–1240. PMID: 24888725.
Article60. Kan M, Koziol-White C, Shumyatcher M, Johnson M, Jester W, Panettieri RA Jr, et al. Airway smooth muscle-specific transcriptomic signatures of glucocorticoid exposure. Am J Respir Cell Mol Biol. 2019; 61:110–120. PMID: 30694689.
Article61. Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001; 29:365–371. PMID: 11726920.
Article62. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. PMID: 11752295.
Article63. Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, et al. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003; 31:68–71. PMID: 12519949.
Article64. Ikeo K, Ishi-i J, Tamura T, Gojobori T, Tateno Y. CIBEX: center for information biology gene expression database. C R Biol. 2003; 326:1079–1082. PMID: 14744116.
Article65. Wang Z, Lachmann A, Ma'ayan A. Mining data and metadata from the gene expression omnibus. Biophys Rev. 2019; 11:103–110. PMID: 30594974.
Article66. Kan M, Shumyatcher M, Diwadkar A, Soliman G, Himes BE. Integration of transcriptomic data identifies global and cell-specific asthma-related gene expression signatures. AMIA Annu Symp Proc. 2018; 2018:1338–1347. PMID: 30815178.67. Kulkarni A, Anderson AG, Merullo DP, Konopka G. Beyond bulk: a review of single cell transcriptomics methodologies and applications. Curr Opin Biotechnol. 2019; 58:129–136. PMID: 30978643.
Article68. Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. 2019; 25:1153–1163. PMID: 31209336.
Article69. Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018; 50:96. PMID: 30089861.