Ann Lab Med.
2020 Mar;40(2):148-154. 10.3343/alm.2020.40.2.148.
Clinical Validity of Next-Generation Sequencing Multi-Gene Panel Testing for Detecting Pathogenic Variants in Patients With Hereditary Breast-Ovarian Cancer Syndrome
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea. microkim@catholic.ac.kr
- 2Catholic Genetic Laboratory Center, Seoul St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- KMID: 2460912
- DOI: http://doi.org/10.3343/alm.2020.40.2.148
Abstract
- BACKGROUND
Hereditary breast and ovarian cancer syndrome (HBOC) is caused by pathogenic variants in BRCA and other cancer-related genes. We analyzed variants in BRCA gene and other cancer-related genes in HBOC patients to evaluate the clinical validity of next-generation sequencing (NGS) multi-gene panel testing.
METHODS
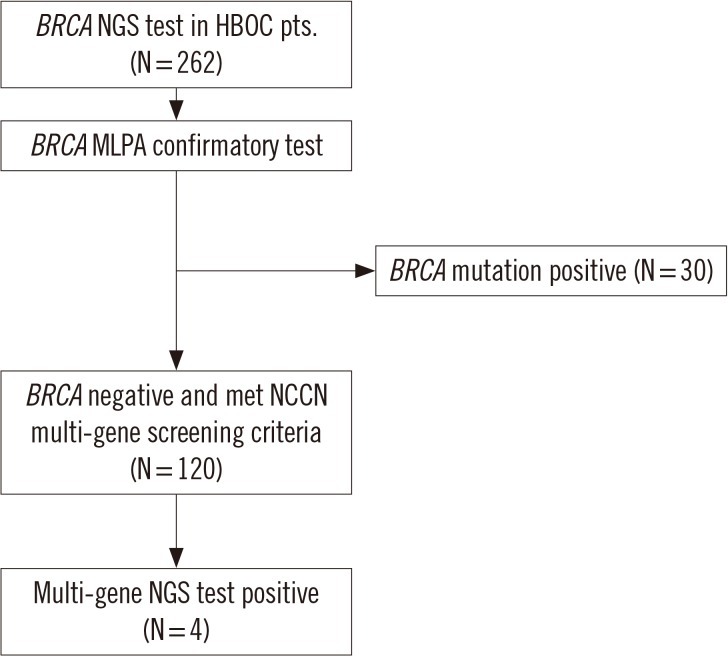
The BRCA1/2 NGS testing was conducted for 262 HBOC patients. Multiplex ligation-dependent probe amplification and direct Sanger sequencing were performed for confirmation. Multi-gene panel testing was conducted for 120 patients who did not possess BRCA1/2 pathogenic variants but met the National Comprehensive Cancer Network criteria.
RESULTS
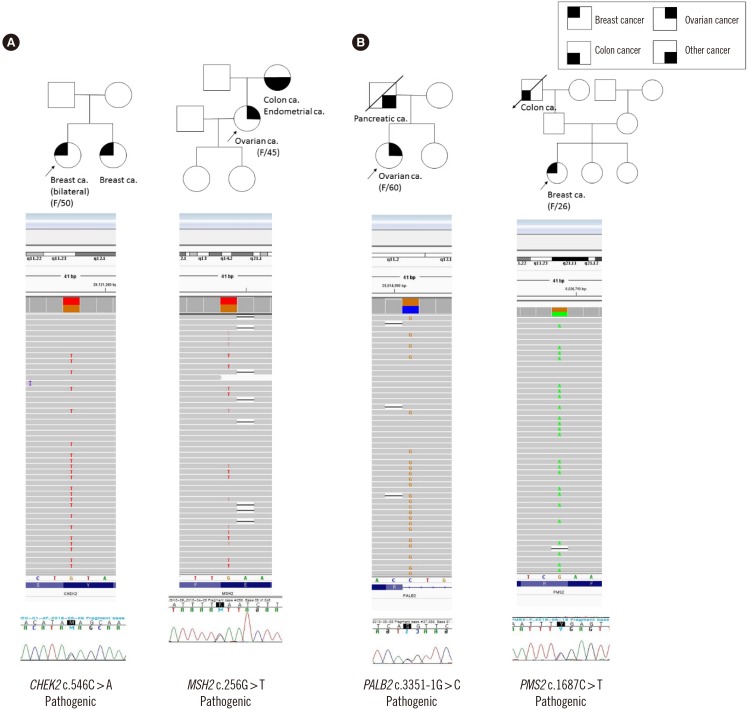
Pathogenic variants in BRCA1/2 were detected in 30 HBOC patients (11.5%). Additionally, four out of the 120 patients possessed pathogenic variants by multi-gene panel testing (3.3%): MSH2 (c.256G>T, p.Glu86*), PMS2 (c.1687C>T, p.Arg563*), CHEK2 (c.546C>A, p.Tyr182*), and PALB2 (c.3351-1G>C). All the four patients had a family history of cancer.
CONCLUSIONS
Multi-gene panel testing could be a significant screening tool for HBOC patients, especially for those with a family history of cancer.
Keyword
MeSH Terms
Figure
Cited by 4 articles
-
From Genetic Testing to Treatment and Prevention of BRCA-Related Breast Cancer
Chang-Seok Ki
Ann Lab Med. 2020;40(2):99-100. doi: 10.3343/alm.2020.40.2.99.Three Cases of False-positive Multiplex Ligation-dependent Probe Amplification of
BRCA1
Kyoung Bo Kim, Sunggyun Park, Jung Sook Ha, Namhee Ryoo, Do-Hoon Kim
Ann Lab Med. 2022;42(4):497-499. doi: 10.3343/alm.2022.42.4.497.Cost-Effectiveness Analysis of Germline and Somatic
BRCA Testing in Patients With Advanced Ovarian Cancer
Jaehyeok Jang, Yoonjung Kim, Jae-Hoon Kim, Sun-Mi Cho, Kyung-A Lee
Ann Lab Med. 2023;43(1):73-81. doi: 10.3343/alm.2023.43.1.73.Cost-effective
BRCA Testing in Advanced Ovarian Cancer
Myungshin Kim
Ann Lab Med. 2023;43(1):3-4. doi: 10.3343/alm.2023.43.1.3.
Reference
-
1. Smith EC. An overview of hereditary breast and ovarian cancer syndrome. J Midwifery Womens Health. 2012; 57:577–584. PMID: 23050669.2. Collaborative Group on Hormonal Factors in Breast Cancer. Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet. 2001; 358:1389–1399. PMID: 11705483.3. Crawford B, Adams SB, Sittler T, van den Akker J, Chan S, Leitner O, et al. Multi-gene panel testing for hereditary cancer predisposition in unsolved high-risk breast and ovarian cancer patients. Breast Cancer Res Treat. 2017; 163:383–390. PMID: 28281021.4. Couch FJ, Nathanson KL, Offit K. Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science. 2014; 343:1466–1470. PMID: 24675953.5. Economopoulou P, Dimitriadis G, Psyrri A. Beyond BRCA: new hereditary breast cancer susceptibility genes. Cancer Treat Rev. 2015; 41:1–8. PMID: 25467110.6. Selkirk CG, Vogel KJ, Newlin AC, Weissman SM, Weiss SM, Wang CH, et al. Cancer genetic testing panels for inherited cancer susceptibility: the clinical experience of a large adult genetics practice. Fam Cancer. 2014; 13:527–536. PMID: 25117502.7. LaDuca H, Stuenkel AJ, Dolinsky JS, Keiles S, Tandy S, Pesaran T, et al. Utilization of multigene panels in hereditary cancer predisposition testing: analysis of more than 2,000 patients. Genet Med. 2014; 16:830–837. PMID: 24763289.8. Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011; 108:18032–18037. PMID: 22006311.9. Desmond A, Kurian AW, Gabree M, Mills MA, Anderson MJ, Kobayashi Y, et al. Clinical actionability of multigene panel testing for hereditary breast and ovarian cancer risk assessment. JAMA Oncol. 2015; 1:943–951. PMID: 26270727.10. Tung N, Battelli C, Allen B, Kaldate R, Bhatnagar S, Bowles K, et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer. 2015; 121:25–33. PMID: 25186627.11. Daly MB, Axilbund JE, Buys S, Crawford B, Farrell CD, Friedman S, et al. Genetic/familial high-risk assessment: breast and ovarian. J Natl Compr Canc Netw. 2010; 8:562–594. PMID: 20495085.12. Kim DH, Chae H, Jo I, Yoo J, Lee H, Jang W, et al. Identification of large genomic rearrangement of BRCA1/2 in high risk patients in Korea. BMC Med Genet. 2017; 18:38. PMID: 28351343.13. Hirotsu Y, Nakagomi H, Sakamoto I, Amemiya K, Oyama T, Mochizuki H, et al. Multigene panel analysis identified germline mutations of DNA repair genes in breast and ovarian cancer. Mol Genet Genomic Med. 2015; 3:459–466. PMID: 26436112.14. Park J, Jang W, Chae H, Kim Y, Chi HY, Kim M. Comparison of targeted next-generation and sanger sequencing for the BRCA1 and BRCA2 mutation screening. Ann Lab Med. 2016; 36:197–201. PMID: 26709275.15. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–424. PMID: 25741868.16. Kang E, Seong MW, Park SK, Lee JW, Lee J, Kim LS, et al. The prevalence and spectrum of BRCA1 and BRCA2 mutations in Korean population: recent update of the Korean Hereditary Breast Cancer (KOHBRA) study. Breast Cancer Res Treat. 2015; 151:157–168. PMID: 25863477.17. Park JS, Lee ST, Nam EJ, Han JW, Lee JY, Kim J, et al. Variants of cancer susceptibility genes in Korean BRCA1/2 mutation-negative patients with high risk for hereditary breast cancer. BMC Cancer. 2018; 18:83. PMID: 29338689.18. Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004; 20:269–276. PMID: 15528792.19. Talseth-Palmer BA, McPhillips M, Groombridge C, Spigelman A, Scott RJ. MSH6 and PMS2 mutation positive Australian Lynch syndrome families: novel mutations, cancer risk and age of diagnosis of colorectal cancer. Hered Cancer Clin Pract. 2010; 8:5. PMID: 20487569.20. Vasovcak P, Krepelova A, Menigatti M, Puchmajerova A, Skapa P, Augustinakova A, et al. Unique mutational profile associated with a loss of TDG expression in the rectal cancer of a patient with a constitutional PMS2 deficiency. DNA Repair (Amst). 2012; 11:616–623. PMID: 22608206.21. Vaughn CP, Robles J, Swensen JJ, Miller CE, Lyon E, Mao R, et al. Clinical analysis of PMS2: mutation detection and avoidance of pseudogenes. Hum Mutat. 2010; 31:588–593. PMID: 20205264.22. Goodenberger ML, Thomas BC, Riegert-Johnson D, Boland CR, Plon SE, Clendenning M, et al. PMS2 monoallelic mutation carriers: the known unknown. Genet Med. 2016; 18:13–19. PMID: 25856668.23. Apostolou P, Papasotiriou I. Current perspectives on CHEK2 mutations in breast cancer. Breast Cancer (Dove Med Press). 2017; 9:331–335. PMID: 28553140.24. Kluska A, Balabas A, Piatkowska M, Czarny K, Paczkowska K, Nowakowska D, et al. PALB2 mutations in BRCA1/2-mutation negative breast and ovarian cancer patients from Poland. BMC Med Genomics. 2017; 10:14. PMID: 28279176.25. Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007; 39:162–164. PMID: 17200671.26. Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006; 22:719–729. PMID: 16793542.27. Harkness EF, Barrow E, Newton K, Green K, Clancy T, Lalloo F, et al. Lynch syndrome caused by MLH1 mutations is associated with an increased risk of breast cancer: a cohort study. J Med Genet. 2015; 52:553–556. PMID: 26101330.28. Scott RJ, McPhillips M, Meldrum CJ, Fitzgerald PE, Adams K, Spigelman AD, et al. Hereditary nonpolyposis colorectal cancer in 95 families: differences and similarities between mutation-positive and mutation-negative kindreds. Am J Hum Genet. 2001; 68:118–127. PMID: 11112663.29. Woo HI, Woo YM, Kim S, Lee ST, Ki CS, Kim JW. Challenges in assessing pathogenicity based on frequency of variants in mismatch repair genes: an extreme case of a MSH2 variant and a meta-analysis. Gene. 2014; 546:421–424. PMID: 24933000.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Utility of Next-Generation Sequencing Panel Including Hereditary Breast and Ovarian Cancer-Related Genes for Pathogenic Variant Detection
- Detection of Germline Mutations in Patients with Epithelial Ovarian Cancer Using Multi-gene Panels: Beyond BRCA1/2
- Clinical Implications of Genetic Testing for Hereditary Breast and Ovarian Cancer Syndrome in the Era of Genomic Medicine: Clinician's Perspectives
- Detection of Germline Mutations in Breast Cancer Patients with Clinical Features of Hereditary Cancer Syndrome Using a Multi-Gene Panel Test
- Diverse genetic spectrum among patients who met the criteria of hereditary breast, ovarian and pancreatic cancer syndrome