Cancer Res Treat.
2018 Jul;50(3):917-925. 10.4143/crt.2017.220.
Detection of Germline Mutations in Patients with Epithelial Ovarian Cancer Using Multi-gene Panels: Beyond BRCA1/2
- Affiliations
-
- 1Hereditary Cancer Clinic, Cancer Prevention Center, Yonsei Cancer Center, Seoul, Korea. NAHMEJ6@yuhs.ac
- 2Department of Obstetrics and Gynecology, Institute of Women's Life Medical Science, Women's Cancer Clinic, Yonsei University College of Medicine, Seoul, Korea.
- 3Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea.
- 4Department of Surgery, Yonsei University College of Medicine, Seoul, Korea.
- 5Department of Medicine, Yonsei University College of Medicine, Seoul, Korea.
- 6Department of Pediatrics, Yonsei University College of Medicine, Seoul, Korea.
- 7Department of Obstetrics and Gynecology, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea.
- KMID: 2417880
- DOI: http://doi.org/10.4143/crt.2017.220
Abstract
- PURPOSE
Next-generation sequencing (NGS) allows simultaneous sequencing of multiple cancer susceptibility genes and may represent a more efficient and less expensive approach than sequential testing. We assessed the frequency of germline mutations in individuals with epithelial ovarian cancer (EOC), using multi-gene panels and NGS.
MATERIALS AND METHODS
Patients with EOC (n=117) with/without a family history of breast or ovarian cancer were recruited consecutively, from March 2016 toDecember 2016.GermlineDNAwas sequenced using 35-gene NGS panel, in order to identify mutations. Upon the detection of a genetic alteration using the panel, results were cross-validated using direct sequencing.
RESULTS
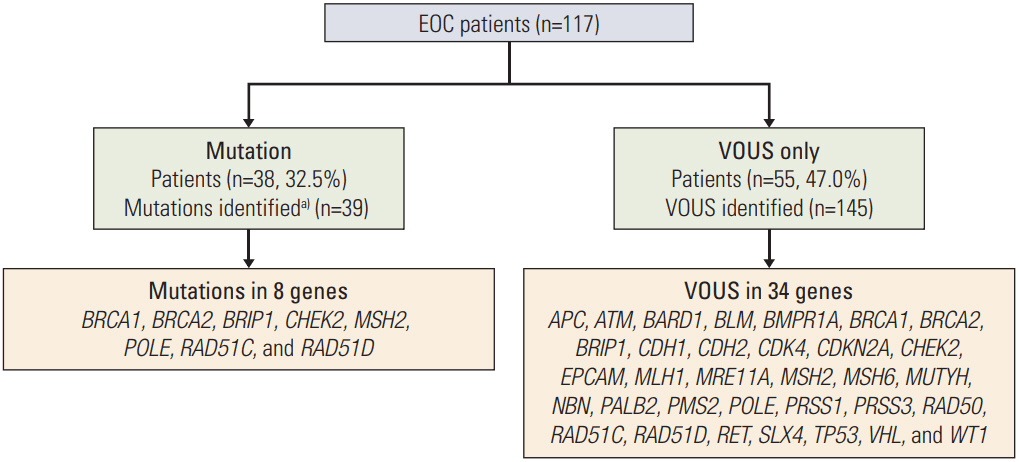
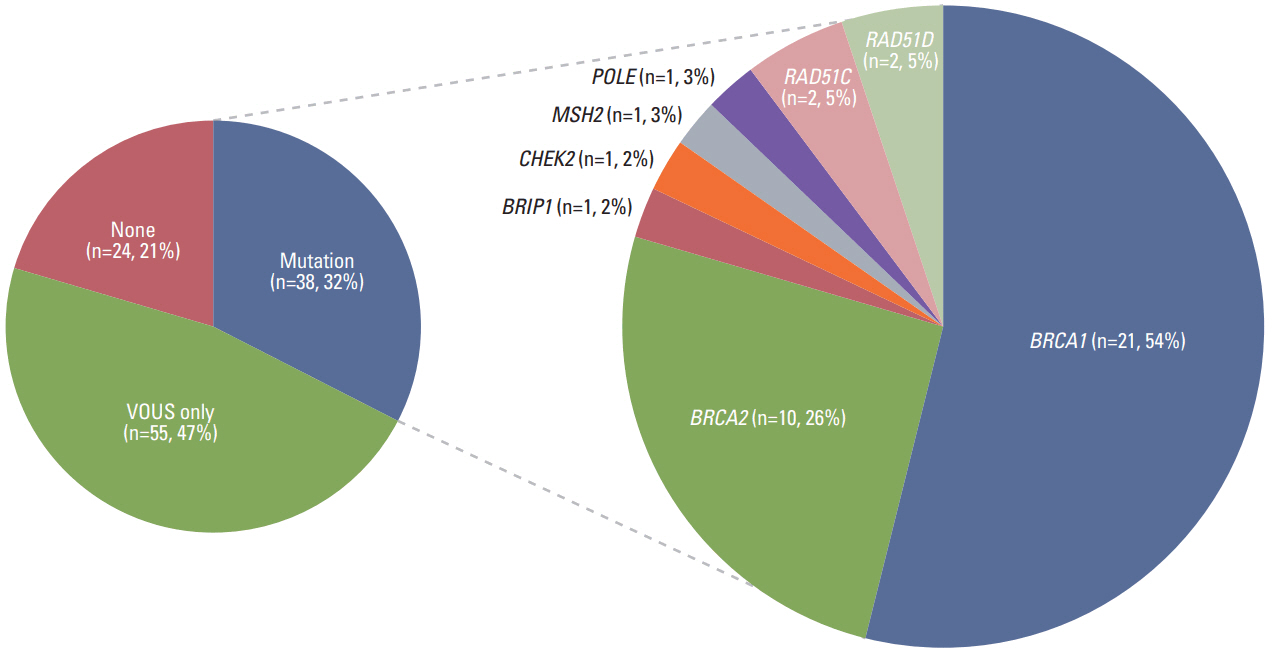
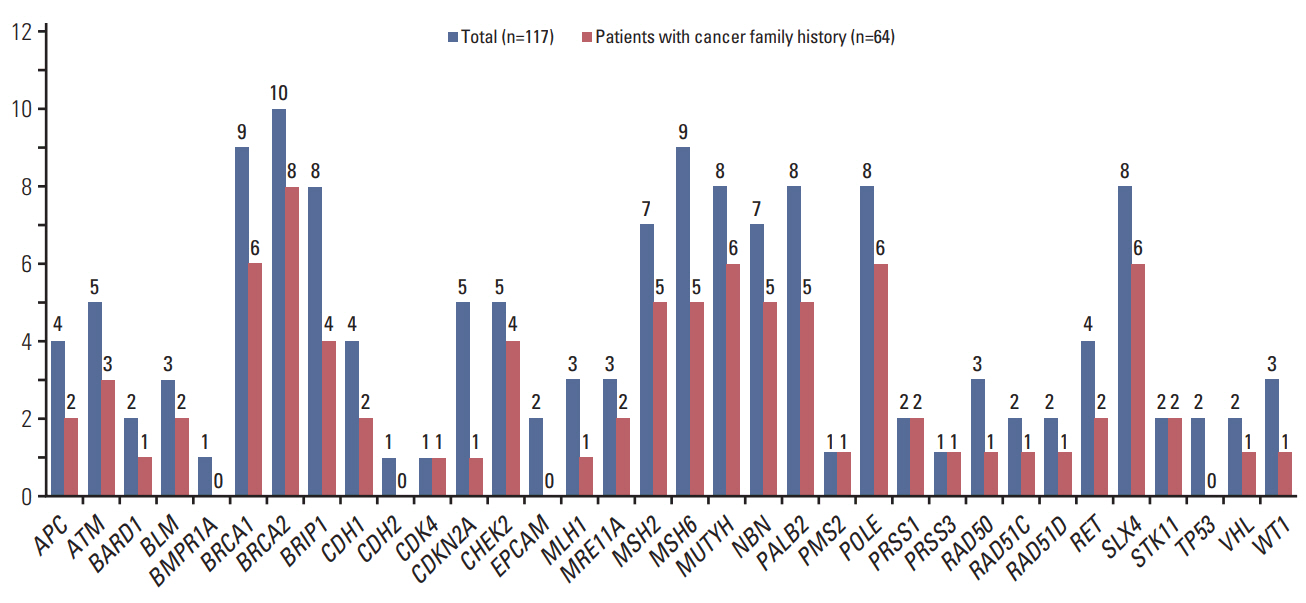
Thirty-eight patients (32.5%) had 39 pathogenic or likely pathogenic mutations in eight genes, including BRCA1 (n=21), BRCA2 (n=10), BRIP1 (n=1), CHEK2 (n=2), MSH2 (n=1), POLE (n=1), RAD51C (n=2), and RAD51D (n=2). Among 64 patients with a family history of cancer, 27 (42.2%) had 27 pathogenic or likely pathogenic mutations, and six (9.3%) had mutations in genes other than BRCA1/2, such as CHECK2, MSH2, POLE, and RAD51C. Fifty-five patients (47.0%) were identified to carry only variants of uncertain significance.
CONCLUSION
Using the multi-gene panel test, we found that, of all patients included in our study, 32.5% had germline cancer-predisposing mutations. NGS was confirmed to substantially improve the detection rates of a wide spectrum of mutations in EOC patients compared with those obtained with the BRCA1/2 testing alone.
Keyword
Figure
Reference
-
References
1. Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007; 25:1329–33.2. van der Kolk DM, de Bock GH, Leegte BK, Schaapveld M, Mourits MJ, de Vries J, et al. Penetrance of breast cancer, ovarian cancer and contralateral breast cancer in BRCA1 and BRCA2 families: high cancer incidence at older age. Breast Cancer Res Treat. 2010; 124:643–51.
Article3. Brose MS, Rebbeck TR, Calzone KA, Stopfer JE, Nathanson KL, Weber BL. Cancer risk estimates for BRCA1 mutation carriers identified in a risk evaluation program. J Natl Cancer Inst. 2002; 94:1365–72.
Article4. Eoh KJ, Park HS, Park JS, Lee ST, Han J, Lee JY, et al. Comparison of clinical outcomes of BRCA1/2 pathologic mutation, variants of unknown significance, or wild type epithelial ovarian cancer patients. Cancer Res Treat. 2017; 49:408–15.
Article5. Domchek SM, Friebel TM, Singer CF, Evans DG, Lynch HT, Isaacs C, et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA. 2010; 304:967–75.6. Finch AP, Lubinski J, Moller P, Singer CF, Karlan B, Senter L, et al. Impact of oophorectomy on cancer incidence and mortality in women with a BRCA1 or BRCA2 mutation. J Clin Oncol. 2014; 32:1547–53.
Article7. Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011; 108:18032–7.
Article8. Frey MK, Kim SH, Bassett RY, Martineau J, Dalton E, Chern JY, et al. Rescreening for genetic mutations using multi-gene panel testing in patients who previously underwent noninformative genetic screening. Gynecol Oncol. 2015; 139:211–5.
Article9. Norquist BM, Harrell MI, Brady MF, Walsh T, Lee MK, Gulsuner S, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016; 2:482–90.
Article10. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–24.
Article11. Pruss D, Morris B, Hughes E, Eggington JM, Esterling L, Robinson BS, et al. Development and validation of a new algorithm for the reclassification of genetic variants identified in the BRCA1 and BRCA2 genes. Breast Cancer Res Treat. 2014; 147:119–32.
Article12. Eggington JM, Bowles KR, Moyes K, Manley S, Esterling L, Sizemore S, et al. A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clin Genet. 2014; 86:229–37.
Article13. De Leeneer K, Van Bockstal M, De Brouwer S, Swietek N, Schietecatte P, Sabbaghian N, et al. Evaluation of RAD51C as cancer susceptibility gene in a large breast-ovarian cancer patient population referred for genetic testing. Breast Cancer Res Treat. 2012; 133:393–8.
Article14. Sopik V, Akbari MR, Narod SA. Genetic testing for RAD51C mutations: in the clinic and community. Clin Genet. 2015; 88:303–12.15. Song H, Dicks E, Ramus SJ, Tyrer JP, Intermaggio MP, Hayward J, et al. Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J Clin Oncol. 2015; 33:2901–7.
Article16. Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005; 37:934–5.
Article17. Rafnar T, Gudbjartsson DF, Sulem P, Jonasdottir A, Sigurdsson A, Jonasdottir A, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet. 2011; 43:1104–7.
Article18. Ramus SJ, Song H, Dicks E, Tyrer JP, Rosenthal AN, Intermaggio MP, et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst. 2015; 107:djv214.
Article19. Cai Z, Chehab NH, Pavletich NP. Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase. Mol Cell. 2009; 35:818–29.
Article20. Weischer M, Bojesen SE, Ellervik C, Tybjaerg-Hansen A, Nordestgaard BG. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol. 2008; 26:542–8.
Article21. Liu Y, Liao J, Xu Y, Chen W, Liu D, Ouyang T, et al. A recurrent CHEK2 p.H371Y mutation is associated with breast cancer risk in Chinese women. Hum Mutat. 2011; 32:1000–3.
Article22. Matulonis UA, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in patients with platinum-sensitive, relapsed serous ovarian cancer and a BRCA mutation: overall survival adjusted for postprogression poly(adenosine diphosphate ribose) polymerase inhibitor therapy. Cancer. 2016; 122:1844–52.23. Matulonis UA, Penson RT, Domchek SM, Kaufman B, Shapira-Frommer R, Audeh MW, et al. Olaparib monotherapy in patients with advanced relapsed ovarian cancer and a germline BRCA1/2 mutation: a multistudy analysis of response rates and safety. Ann Oncol. 2016; 27:1013–9.
Article24. NCCN clinical practice guidelines in oncology: genetic/familial high risk assessment: breast and ovarian (version 1.2017). Fort Washington, PA: National Comprehensive Cancer Network;2016.25. Miesfeldt S, Lamb A, Duarte C. Management of genetic syndromes predisposing to gynecologic cancers. Curr Treat Options Oncol. 2013; 14:34–50.
Article26. Kurian AW, Hare EE, Mills MA, Kingham KE, McPherson L, Whittemore AS, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014; 32:2001–9.
Article27. Elsayed FA, Kets CM, Ruano D, van den Akker B, Mensenkamp AR, Schrumpf M, et al. Germline variants in POLE are associated with early onset mismatch repair deficient colorectal cancer. Eur J Hum Genet. 2015; 23:1080–4.
Article28. Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002; 20:1480–90.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Germline Mutations and polymorphisms of BRCA1 and BRCA2 in Sporadic Ovarian Carcinoma
- A Study for Germline Mutation of BRCA1 in Early Onset Breast Cancer Patients
- Frequency of BRCA1 and BRCA2 Germline Mutations Detected by Protein Truncation Test and Cumulative Risks of Breast and Ovarian Cancer among Mutation Carriers in Japanese Breast Cancer Families
- Prevalence of germline BRCA mutations among women with carcinoma of the peritoneum or fallopian tube
- Germline Mutations of BRCA1 Gene in Korean Breast and/or Ovarian Cancer Families