Korean J Radiol.
2019 Oct;20(10):1431-1440. 10.3348/kjr.2019.0212.
Deep Learning Algorithm for Reducing CT Slice Thickness: Effect on Reproducibility of Radiomic Features in Lung Cancer
- Affiliations
-
- 1Department of Radiology and Research Institute of Radiology, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea. sangmin.lee.md@gmail.com
- 2Department of Convergence Medicine, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea.
- 3VUNO Inc., Seoul, Korea.
- KMID: 2459162
- DOI: http://doi.org/10.3348/kjr.2019.0212
Abstract
OBJECTIVE
To retrospectively assess the effect of CT slice thickness on the reproducibility of radiomic features (RFs) of lung cancer, and to investigate whether convolutional neural network (CNN)-based super-resolution (SR) algorithms can improve the reproducibility of RFs obtained from images with different slice thicknesses.
MATERIALS AND METHODS
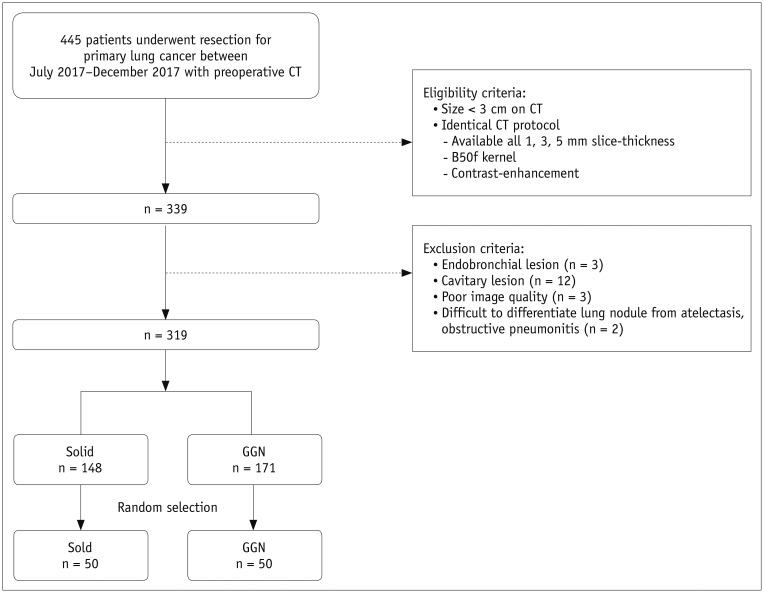
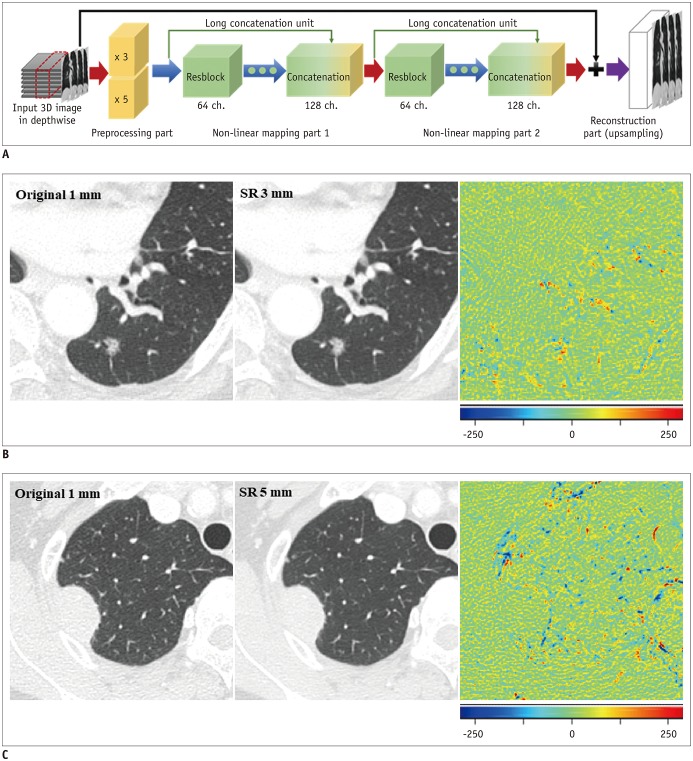
CT images with 1-, 3-, and 5-mm slice thicknesses obtained from 100 pathologically proven lung cancers between July 2017 and December 2017 were evaluated. CNN-based SR algorithms using residual learning were developed to convert thick-slice images into 1-mm slices. Lung cancers were semi-automatically segmented and a total of 702 RFs (tumor intensity, texture, and wavelet features) were extracted from 1-, 3-, and 5-mm slices, as well as the 1-mm slices generated from the 3- and 5-mm images. The stabilities of the RFs were evaluated using concordance correlation coefficients (CCCs).
RESULTS
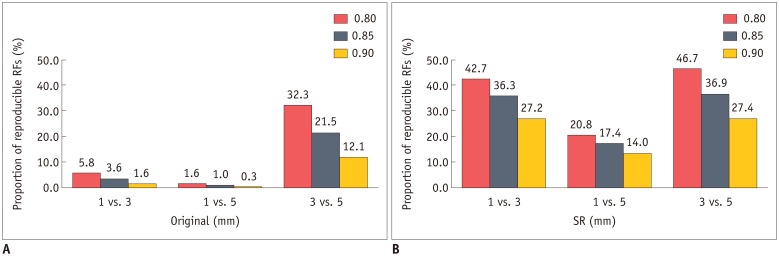
The mean CCCs for the comparisons of original 1 mm vs. 3 mm, 1 mm vs. 5 mm, and 3 mm vs. 5 mm images were 0.41, 0.27, and 0.65, respectively (p < 0.001 for all comparisons). Tumor intensity features showed the best reproducibility while wavelets showed the lowest reproducibility. The majority of RFs failed to achieve reproducibility (CCC ≥ 0.85; 3.6%, 1.0%, and 21.5%, respectively). After applying the CNN-based SR algorithms, the reproducibility significantly improved in all three pairings (mean CCCs: 0.58, 0.45, and 0.72; p < 0.001 for all comparisons). The reproducible RFs also increased (36.3%, 17.4%, and 36.9%, respectively).
CONCLUSION
The reproducibility of RFs in lung cancer is significantly influenced by CT slice thickness, which can be improved by the CNN-based SR algorithms.
Keyword
Figure
Reference
-
1. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016; 278:563–577. PMID: 26579733.
Article2. Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014; 5:4006. PMID: 24892406.
Article3. Zhang B, Tian J, Dong D, Gu D, Dong Y, Zhang L, et al. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res. 2017; 23:4259–4269. PMID: 28280088.
Article4. Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology. 2016; 281:947–957. PMID: 27347764.
Article5. Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, Castro-García M, Villas MV, Mansilla Legorburo F, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018; 288:407–415. PMID: 29688159.6. He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep. 2016; 6:34921. PMID: 27721474.
Article7. Kim J, Lee JK, Lee KM. Accurate image super-resolution using very deep convolutional networks. In : The IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 June 27–30; Las Vegas, NV, USA. p. 1646–1654.8. Bhavsar A, Wu G, Lian J, Shen D. Resolution enhancement of lung 4D-CT via group-sparsity. Med Phys. 2013; 40:121717. PMID: 24320503.
Article9. Rueda A, Malpica N, Romero E. Single-image super-resolution of brain MR images using overcomplete dictionaries. Med Image Anal. 2013; 17:113–132. PMID: 23102924.
Article10. Coupé P, Manjón JV, Chamberland M, Descoteaux M, Hiba B. Collaborative patch-based super-resolution for diffusion-weighted images. Neuroimage. 2013; 83:245–261. PMID: 23791914.
Article11. Bahrami K, Shi F, Rekik I, Gao Y, Shen D. 7T-guided super-resolution of 3T MRI. Med Phys. 2017; 44:1661–1677. PMID: 28177548.
Article12. Kim C, Lee SM, Choe J, Chae EJ, Do KH, Seo JB. Volume doubling time of lung cancer detected in idiopathic interstitial pneumonia: comparison with that in chronic obstructive pulmonary disease. Eur Radiol. 2018; 28:1402–1409. PMID: 29038933.
Article13. Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, et al. Robust Radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One. 2014; 9:e102107. PMID: 25025374.
Article14. Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989; 45:255–268. PMID: 2720055.
Article15. Zhao B, Tan Y, Tsai WY, Qi J, Xie C, Lu L, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016; 6:23428. PMID: 27009765.
Article16. Larue RTHM, van Timmeren JE, de Jong EEC, Feliciani G, Leijenaar RTH, Schreurs WMJ, et al. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol. 2017; 56:1544–1553. PMID: 28885084.
Article17. Lu L, Ehmke RC, Schwartz LH, Zhao B. Assessing agreement between radiomic features computed for multiple CT imaging settings. PLoS One. 2016; 11:e0166550. PMID: 28033372.
Article18. Bankier AA, MacMahon H, Goo JM, Rubin GD, Schaefer-Prokop CM, Naidich DP. Recommendations for measuring pulmonary nodules at CT: a statement from the Fleischner Society. Radiology. 2017; 285:584–600. PMID: 28650738.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Segmentation of lung fields from chest radiographs-a radiomic feature-based approach
- Detectability of Various Sizes of Honeycombing Cysts in an Inflated and Fixed Lung Specimen: the Effect of CT Section Thickness
- Effect of the slice thickness and the size of region of interest on CT number
- Assessment of a Deep Learning Algorithm for the Detection of Rib Fractures on Whole-Body Trauma Computed Tomography
- Application of Artificial Intelligence in Lung Cancer Screening