Brain Tumor Res Treat.
2019 Apr;7(1):33-38. 10.14791/btrt.2019.7.e20.
von Willebrand Factor Gene Expression in Primary Lower Grade Glioma: Mutually Co-Occurring Mutations in von Willebrand Factor, ATRX, and TP53
- Affiliations
-
- 1Department of Radiation Oncology, Icahn School of Medicine at Mount Sinai, New York, NY, USA. steven.lehrer@mssm.edu
- 2Severn Health Solutions, Severna Park, MD, USA.
- KMID: 2444793
- DOI: http://doi.org/10.14791/btrt.2019.7.e20
Abstract
- BACKGROUND
Venous thromboembolism is a common complication in patients with glioma. The clotting factor von Willebrand factor (VWF) is a highly adhesive procoagulant molecule that mediates platelet adhesion to endothelial and subendothelial surfaces. In the current analysis, we examined The Cancer Genome Atlas (TCGA) data to assess the VWF gene in patients with lower grade gliomas.
METHODS
For newly diagnosed gliomas, we evaluated the association between VWF and overall survival in the Genomic Data Commons TCGA Lower Grade Glioma (LGG) dataset in TCGA. Simple statistics were calculated to identify patterns of mutual exclusivity or co-occurrence of VWF mutations. For each pair of query genes an odds ratio was calculated that indicates the likelihood that the mutations in the two genes are mutually exclusive or co-occurrent across the selected cases. To determine whether the identified relationship was significant for a gene pair, Fisher's exact test was performed.
RESULTS
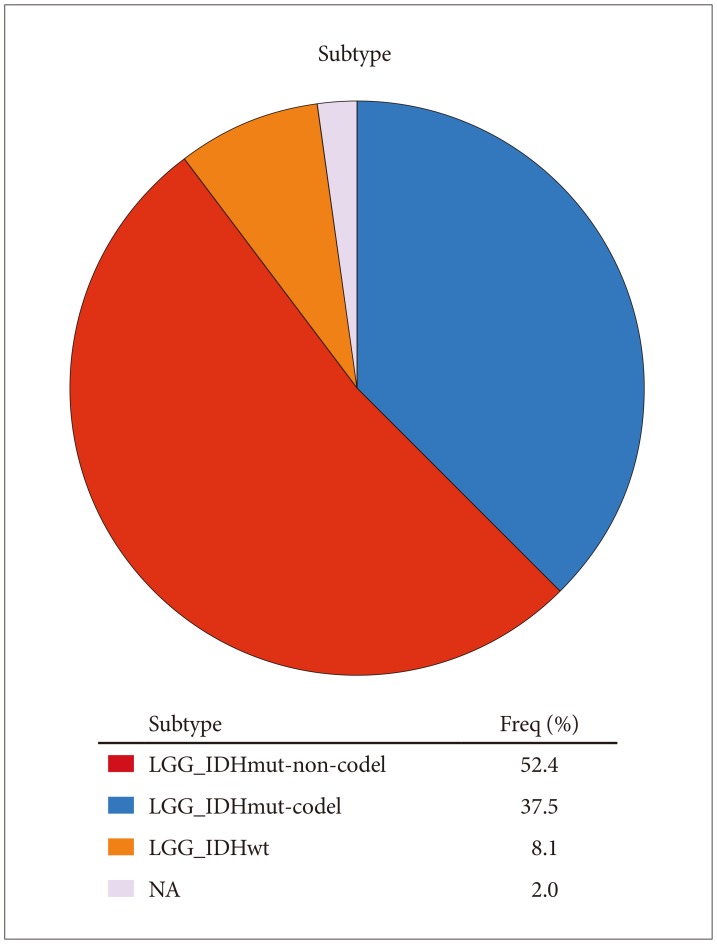
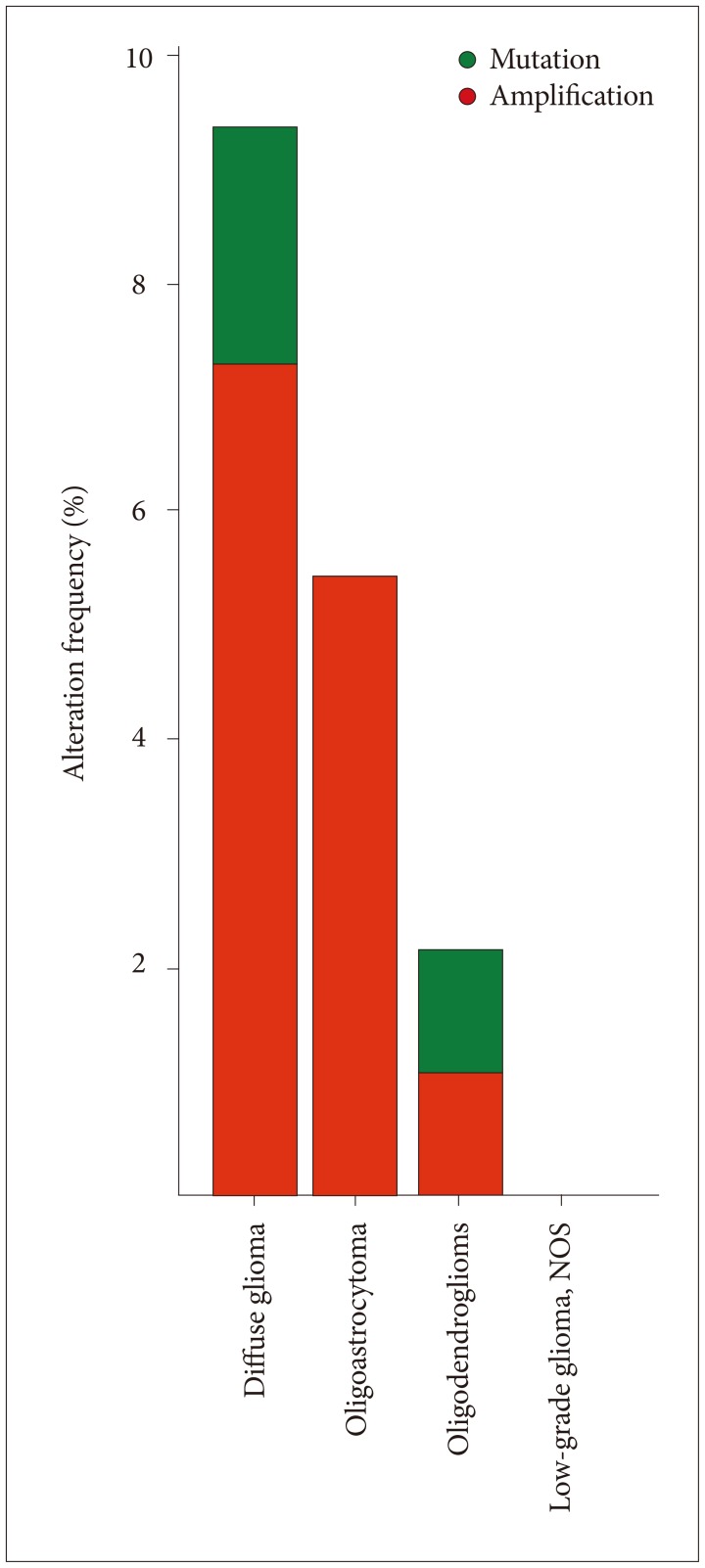
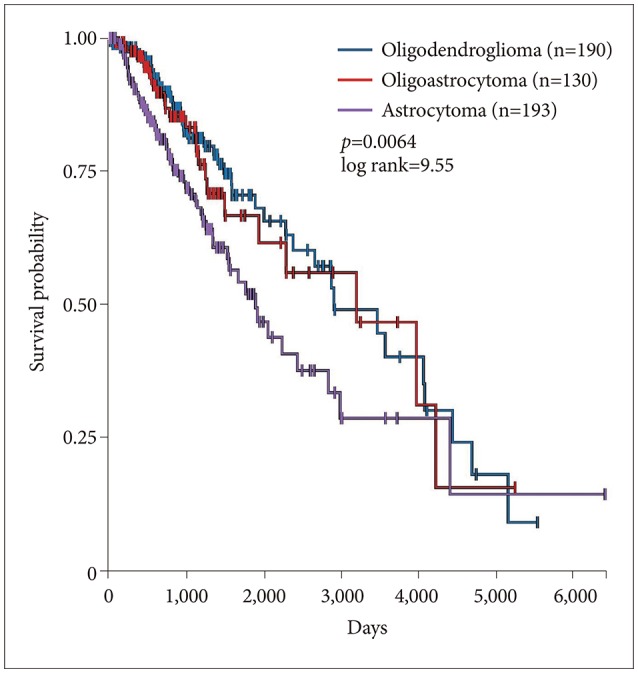
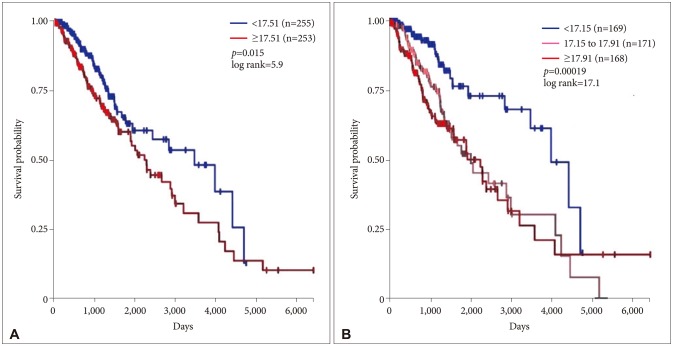
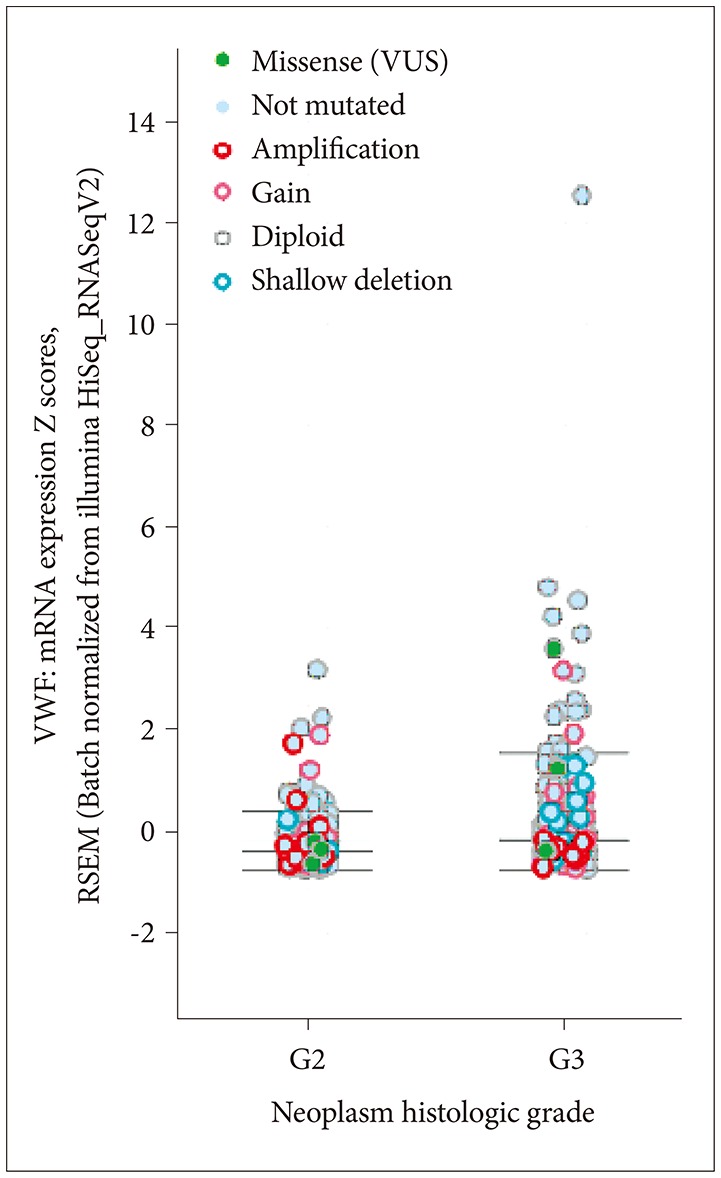
Lower grade gliomas with less VWF gene expression had significantly better survival than those with more VWF gene expression (hazard ratio 0.64, 95% confidence interval 0.44 to 0.92, p=0.015 log rank test). When we analyzed the data with Cox regression, VWF expression had a significant effect on survival (p=0.02) that was unrelated to the effect of IDH1 expression (p=0.062), TP53 expression (p=0.135), independent of ATRX expression (p=0.021) and histology (astrocytoma versus oligoastrocytoma and oligodendroglioma, p=0.002). VWF mutations significantly co-occur with mutations in TP53 and ATRX (p<0.001).
CONCLUSION
The deleterious prognostic effect of VWF expression and its co-occurrent mutations with TP53 and ATRX in lower grade gliomas are not surprising, given VWF's role in other cancers. Therefore, VWF gene expression may be a clinically important risk marker in lower grade glioma.
MeSH Terms
Figure
Reference
-
1. Norden AD, Bartolomeo J, Tanaka S, et al. Safety of concurrent bevacizumab therapy and anticoagulation in glioma patients. J Neurooncol. 2012; 106:121–125. PMID: 21706358.
Article2. Al Megren M, De Wit C, Al Qahtani M, Le Gal G, Carrier M. Management of venous thromboembolism in patients with glioma. Thromb Res. 2017; 156:105–108. PMID: 28624717.
Article3. Simanek R, Vormittag R, Hassler M, et al. Venous thromboembolism and survival in patients with high-grade glioma. Neuro Oncol. 2007; 9:89–95. PMID: 17327573.
Article4. Hua Y, Tang L, Keep RF, et al. The role of thrombin in gliomas. J Thromb Haemost. 2005; 3:1917–1923. PMID: 15975137.
Article5. Mojiri A, Stoletov K, Carrillo MA, et al. Functional assessment of von Willebrand factor expression by cancer cells of non-endothelial origin. Oncotarget. 2017; 8:13015–13029. PMID: 28035064.
Article6. Marfia G, Navone SE, Fanizzi C, et al. Prognostic value of preoperative von Willebrand factor plasma levels in patients with glioblastoma. Cancer Med. 2016; 5:1783–1790. PMID: 27236861.
Article7. R2 Support Team. Working with Kaplan Meier. R2 Tutorials. Release 3.1.2. Amsterdam: Jan Koster (Academic Medical Center);2017. p. 73–82.8. Shahriyari L. Effect of normalization methods on the performance of supervised learning algorithms applied to HTSeq-FPKM-UQ data sets: 7SK RNA expression as a predictor of survival in patients with colon adenocarcinoma. Brief Bioinform. 2017; 11. 03. DOI: 10.1093/bib/bbx153.
Article9. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013; 6:pl1. PMID: 23550210.
Article10. Ceccarelli M, Barthel FP, Malta TM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016; 164:550–563. PMID: 26824661.11. Wang S, Jin F, Fan W, et al. Gene expression meta-analysis in diffuse low-grade glioma and the corresponding histological subtypes. Sci Rep. 2017; 7:11741. PMID: 28924174.
Article12. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016; 131:803–820. PMID: 27157931.
Article14. Venneti S, Huse JT. The evolving molecular genetics of low-grade glioma. Adv Anat Pathol. 2015; 22:94–101. PMID: 25664944.
Article15. Buckner J, Giannini C, Eckel-Passow J, et al. Management of diffuse lowgrade gliomas in adults - use of molecular diagnostics. Nat Rev Neurol. 2017; 13:340–351. PMID: 28497806.
Article16. Cancer Genome Atlas Research Network. Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015; 372:2481–2498. PMID: 26061751.17. Claus EB, Walsh KM, Wiencke JK, et al. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus. 2015; 38:E6.
Article18. Stupp R, Janzer RC, Hegi ME, Villemure JG, Mirimanoff RO. Prognostic factors for low-grade gliomas. Semin Oncol. 2003; 30(6 Suppl 19):23–28.
Article19. Furtner J, Bender B, Braun C, et al. Prognostic value of blood flow measurements using arterial spin labeling in gliomas. PLoS One. 2014; 9:e99616. PMID: 24911025.
Article20. Lehrer S, Dembitzer FR, Rheinstein PH, Rosenzweig KE. In primary glioblastoma fewer tumor copy number segments of the F13A1 gene are associated with poorer survival. Thromb Res. 2018; 167:12–14. PMID: 29753279.
Article21. Varki A. Trousseau’s syndrome: multiple definitions and multiple mechanisms. Blood. 2007; 110:1723–1729. PMID: 17496204.
Article22. Bauer AT, Suckau J, Frank K, et al. von Willebrand factor fibers promote cancer-associated platelet aggregation in malignant melanoma of mice and humans. Blood. 2015; 125:3153–3163. PMID: 25977583.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Laboratory assessment of von Willebrand factor for classification of von Willebrand disease
- Detection of Novel C4517G (Ser743Trp) Mutation in a Family with Type 2A von Willebrand Disease
- Three cases of type I von Willebrand disease in a family
- Investigation of von Willebrand Factor Gene Mutations in Korean von Willebrand Disease Patients
- Effect of immune-mediated vascular injury on the coagulation- regulatory mechanism of the human endothelial cells; changes of tissue-type plasminogen activator, plasminogen activator inhibitor- 1 and von Willebrand factor