Investig Clin Urol.
2016 Jun;57(Suppl 1):S52-S59. 10.4111/icu.2016.57.S1.S52.
Aberrantly expressed microRNAs in the context of bladder tumorigenesis
- Affiliations
-
- 1Theragen Etex Bio Institute, Suwon, Korea. jasonsjkim@therabio.kr
- 2Department of Urology, Chungbuk National University College of Medicine, Cheongju, Korea.
- KMID: 2426517
- DOI: http://doi.org/10.4111/icu.2016.57.S1.S52
Abstract
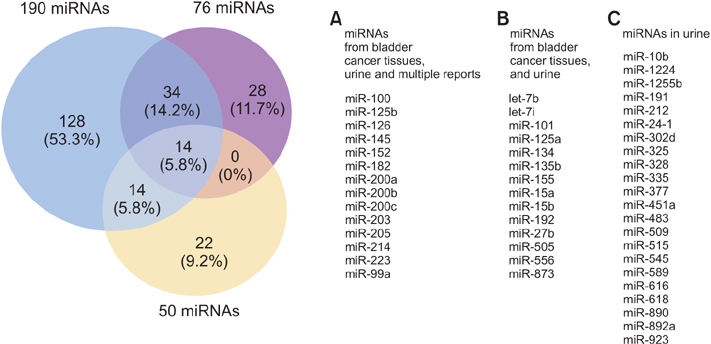
- MicroRNAs (miRNAs), small noncoding RNAs 19-22 nucleotides in length, play a major role in negative regulation of gene expression at the posttranscriptional level. Several miRNAs act as tumor suppressors or oncogenes that control cell differentiation, proliferation, apoptosis, or angiogenesis during tumorigenesis. To date, 19 research groups have published large-scale expression profiles that identified 261 miRNAs differentially expressed in bladder cancer, of which 76 were confirmed to have consistent expression patterns by two or more groups. These consistently expressed miRNAs participated in regulation of multiple biological processes and factors, including axon guidance, cancer-associated proteoglycans, and the ErbB and transforming growth factorbeta signaling pathways. Because miRNAs can be released from cancer cells into urine via secreted particles, we propose that miRNAs differentially expressed between tissue and urine could serve as predictors of bladder cancer, and could thus be exploited for noninvasive diagnosis.
Keyword
MeSH Terms
Figure
Reference
-
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012; 62:10–29.2. Kim SM, Kang HW, Kim WT, Kim YJ, Yun SJ, Lee SC, et al. Cell-free microRNA-214 from urine as a biomarker for nonmuscle-invasive bladder cancer. Korean J Urol. 2013; 54:791–796.3. Messing E. Markers of detection. Urol Oncol. 2007; 25:344–347.4. Quan C, Cha EJ, Lee HL, Han KH, Lee KM, Kim WJ. Enhanced expression of peroxiredoxin I and VI correlates with development, recurrence and progression of human bladder cancer. J Urol. 2006; 175:1512–1516.5. Babjuk M, Oosterlinck W, Sylvester R, Kaasinen E, Bohle A, Palou-Redorta J, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder. Eur Urol. 2008; 54:303–314.6. Sylvester RJ. Natural history, recurrence, and progression in superficial bladder cancer. ScientificWorldJournal. 2006; 6:2617–2625.7. Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009; 374:239–249.8. Knowles MA. Role of FGFR3 in urothelial cell carcinoma: biomarker and potential therapeutic target. World J Urol. 2007; 25:581–593.9. Pandith AA, Shah ZA, Siddiqi MA. Oncogenic role of fibroblast growth factor receptor 3 in tumorigenesis of urinary bladder cancer. Urol Oncol. 2013; 31:398–406.10. Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005; 5:713–725.11. Bakkar AA, Wallerand H, Radvanyi F, Lahaye JB, Pissard S, Lecerf L, et al. FGFR3 and TP53 gene mutations define two distinct pathways in urothelial cell carcinoma of the bladder. Cancer Res. 2003; 63:8108–8112.12. Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999; 23:18–20.13. Wallerand H, Reiter RR, Ravaud A. Molecular targeting in the treatment of either advanced or metastatic bladder cancer or both according to the signalling pathways. Curr Opin Urol. 2008; 18:524–532.14. Cazier JB, Rao SR, McLean CM, Walker AK, Wright BJ, Jaeger EE, et al. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nat Commun. 2014; 5:3756.15. Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3' untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009; 16:144–150.16. Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004; 5:396–400.17. Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006; 94:776–780.18. Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs: the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009; 9:293–302.19. Bandres E, Agirre X, Ramirez N, Zarate R, Garcia-Foncillas J. MicroRNAs as cancer players: potential clinical and biological effects. DNA Cell Biol. 2007; 26:273–282.20. Catto JW, Miah S, Owen HC, Bryant H, Myers K, Dudziec E, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009; 69:8472–8481.21. Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007; 25:387–392.22. Hidaka H, Seki N, Yoshino H, Yamasaki T, Yamada Y, Nohata N, et al. Tumor suppressive microRNA-1285 regulates novel molecular targets: aberrant expression and functional significance in renal cell carcinoma. Oncotarget. 2012; 3:44–57.23. Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, et al. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer. 2013; 132:2479–2491.24. Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol. 2013; 10:396–404.25. Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008; 18:1509–1517.26. Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010; 464:773–777.27. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008; 5:621–628.28. Mutz KO, Heilkenbrinker A, Lonne M, Walter JG, Stahl F. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol. 2013; 24:22–30.29. Nagalakshmi U, Waern K, Snyder M. RNA-Seq: a method for comprehensive transcriptome analysis. Curr Protoc Mol Biol. 2010; Chapter 4:Unit 4.11.1-13.30. Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011; 8:469–477.31. Bottomly D, Walter NA, Hunter JE, Darakjian P, Kawane S, Buck KJ, et al. Evaluating gene expression in C57BL/6J and DBA/2J mouse striatum using RNA-Seq and microarrays. PLoS One. 2011; 6:e17820.32. Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlen M, et al. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res. 2012; 40:10084–10097.33. Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014; 9:e78644.34. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079.35. Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012; 40:37–52.36. Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009; 69:2623–2629.37. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012; 9:357–359.38. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015; 4:e05005.39. Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016; 44:D239–D247.40. Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015; 43:W460–W466.41. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004; 10:1507–1517.42. Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013; 41:W169–W173.43. Lin T, Dong W, Huang J, Pan Q, Fan X, Zhang C, et al. MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol. 2009; 181:1372–1380.44. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005; 435:834–838.45. Canturk KM, Ozdemir M, Can C, Oner S, Emre R, Aslan H, et al. Investigation of key miRNAs and target genes in bladder cancer using miRNA profiling and bioinformatic tools. Mol Biol Rep. 2014; 41:8127–8135.46. Han Y, Chen J, Zhao X, Liang C, Wang Y, Sun L, et al. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One. 2011; 6:e18286.47. Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, et al. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS One. 2014; 9:e84311.48. Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, et al. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011; 104:808–818.49. Song T, Xia W, Shao N, Zhang X, Wang C, Wu Y, et al. Differential miRNA expression profiles in bladder urothelial carcinomas. Asian Pac J Cancer Prev. 2010; 11:905–911.50. Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009; 125:345–352.51. Ratert N, Meyer HA, Jung M, Lioudmer P, Mollenkopf HJ, Wagner I, et al. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J Mol Diagn. 2013; 15:695–705.52. Chen YH, Wang SQ, Wu XL, Shen M, Chen ZG, Chen XG, et al. Characterization of microRNAs expression profiling in one group of Chinese urothelial cell carcinoma identified by Solexa sequencing. Urol Oncol. 2013; 31:219–227.53. Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009; 69:4851–4860.54. Zhu J, Jiang Z, Gao F, Hu X, Zhou L, Chen J, et al. A systematic analysis on DNA methylation and the expression of both mRNA and microRNA in bladder cancer. PLoS One. 2011; 6:e28223.55. Li X, Chen J, Hu X, Huang Y, Li Z, Zhou L, et al. Comparative mRNA and microRNA expression profiling of three genitourinary cancers reveals common hallmarks and cancer-specific molecular events. PLoS One. 2011; 6:e22570.56. Wang G, Zhang H, He H, Tong W, Wang B, Liao G, et al. Upregulation of microRNA in bladder tumor tissue is not common. Int Urol Nephrol. 2010; 42:95–102.57. Jia AY, Castillo-Martin M, Bonal DM, Sanchez-Carbayo M, Silva JM, Cordon-Cardo C. MicroRNA-126 inhibits invasion in bladder cancer via regulation of ADAM9. Br J Cancer. 2014; 110:2945–2954.58. Cheng Y, Deng X, Yang X, Li P, Zhang X, Li P, et al. Urine microRNAs as biomarkers for bladder cancer: a diagnostic meta-analysis. Onco Targets Ther. 2015; 8:2089–2096.59. Tolle A, Jung M, Rabenhorst S, Kilic E, Jung K, Weikert S. Identification of microRNAs in blood and urine as tumour markers for the detection of urinary bladder cancer. Oncol Rep. 2013; 30:1949–1956.60. Wang G, Chan ES, Kwan BC, Li PK, Yip SK, Szeto CC, et al. Expression of microRNAs in the urine of patients with bladder cancer. Clin Genitourin Cancer. 2012; 10:106–113.61. Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010; 28:655–661.62. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010; 56:1733–1741.63. Miah S, Dudziec E, Drayton RM, Zlotta AR, Morgan SL, Rosario DJ, et al. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br J Cancer. 2012; 107:123–128.64. Yun SJ, Jeong P, Kim WT, Kim TH, Lee YS, Song PH, et al. Cell-free microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. Int J Oncol. 2012; 41:1871–1878.65. Snowdon J, Boag S, Feilotter H, Izard J, Siemens DR. A pilot study of urinary microRNA as a biomarker for urothelial cancer. Can Urol Assoc J. 2013; 7:28–32.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Aspect of MicroRNA in Lung Cancer
- The Molecular Pathogenesis of Pituitary Adenomas: An Update
- MicroRNA Expression Pattern in Intraductal Papillary Mucinous Neoplasm
- Long non-coding RNA linc00152 acting as a promising oncogene in cancer progression
- Amplication of C-myc and rearrangement of erb B2 in bladder cancer