J Vet Sci.
2017 Sep;18(3):317-326. 10.4142/jvs.2017.18.3.317.
Genetic diversity and phylogenetic analysis of porcine reproductive and respiratory syndrome virus in southern China from 2007 to 2014
- Affiliations
-
- 1College of Animal Science, South China Agricultural University, Guangzhou 510642, China. majy2400@scau.edu.cn
- 2Henan University of Science and Technology, Luoyang 471000, China.
- 3Guangdong Wen's Foodstuff Group Co., Ltd., Yunfu 527400, China.
- KMID: 2412449
- DOI: http://doi.org/10.4142/jvs.2017.18.3.317
Abstract
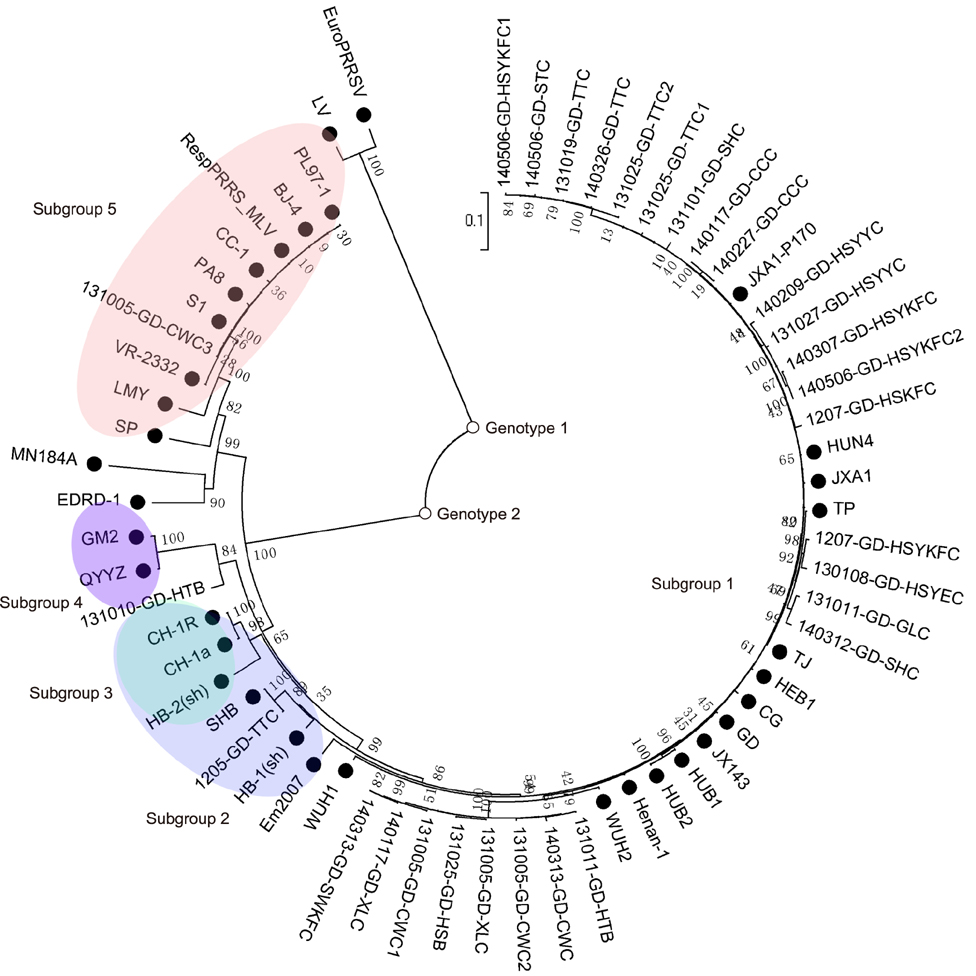
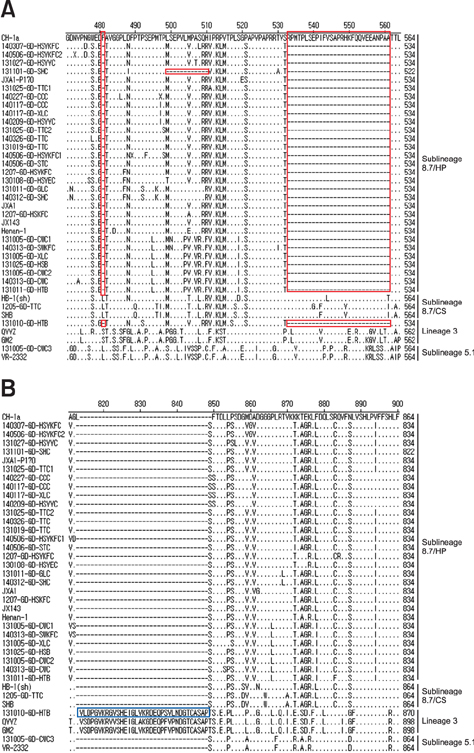
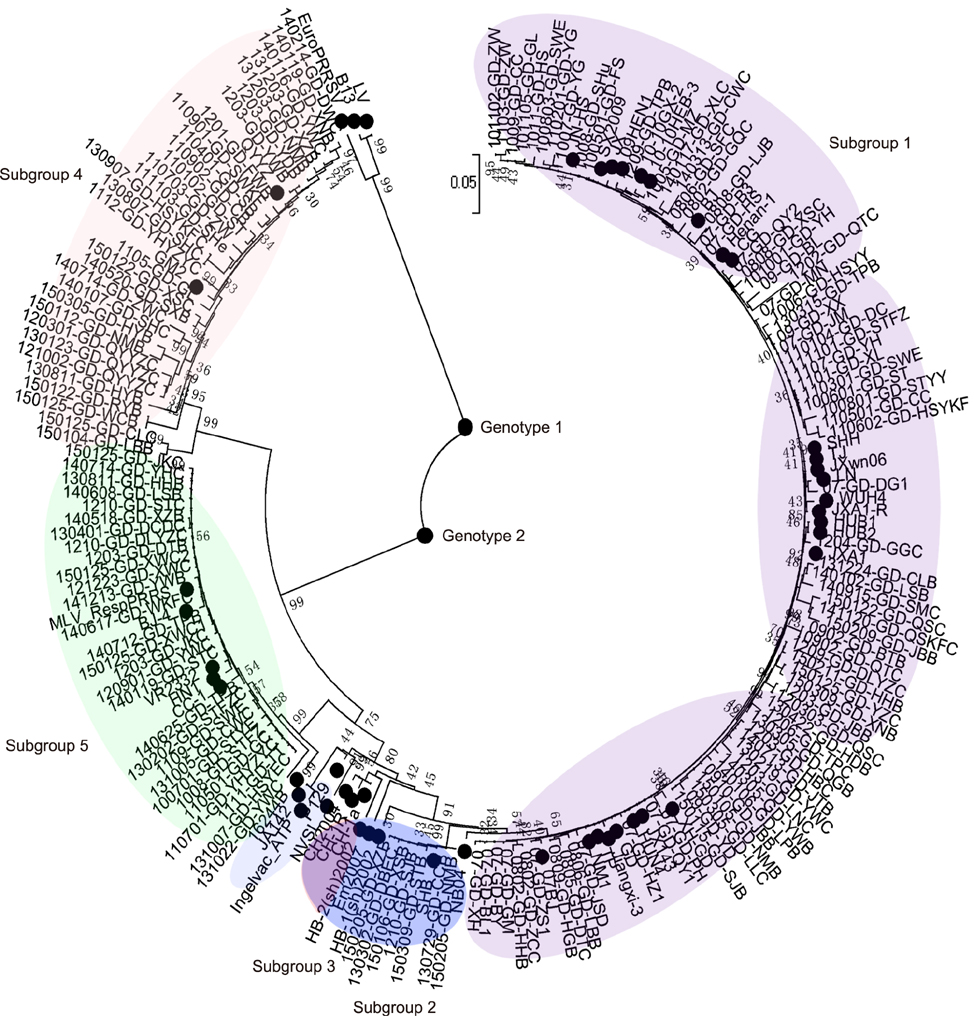
- Porcine reproductive and respiratory syndrome virus (PRRSV) has a high degree of genetic variation. In this study, we characterized the genetic variation and evolutionary relationships among circulating PRRSV strains in southern China. We analyzed 29 NSP2 strains and 150 ORF5 strains from clinical samples collected in southern China during 2007-2014. The alignment results showed that the nucleotide identity similarities of the two genes among these strains were 80.5%-99.7% and 80.9%-100%, respectively. Phylogenetic analysis based on the NSP2 gene showed that highly pathogenic (HP)-PRRSV was still the dominant virus in southern China from 2013 to 2014. Compared with reference strains CH-1a and VR-2332, the field strain 131101-GD-SHC, which shared high homology with JXA1-P170, had a novel 12 amino acid deletion at position 499-510. Phylogenetic analysis based on the ORF5 gene showed that HP-PRRSV, VR2332-like strains, and QYYZ-like strains were simultaneously circulating in southern China from 2007 to 2014, suggesting that, in recent years, the type 2 PRRSV was more diverse in southern China. In conclusion, mutations in the decoy epitope and primary neutralizing epitope could be markers of viral evolution and used to study evolutionary relationships among PRRSV strains in China.
MeSH Terms
-
Animals
China/epidemiology
Genetic Variation/*genetics
Phylogeny
Polymerase Chain Reaction/veterinary
Porcine Reproductive and Respiratory Syndrome/epidemiology/virology
Porcine respiratory and reproductive syndrome virus/*genetics
Sequence Alignment/veterinary
Sequence Analysis, DNA/veterinary
Sequence Analysis, Protein/veterinary
Swine
Figure
Reference
-
1. An TQ, Tian ZJ, Xiao Y, Li R, Peng JM, Wei TC, Zhang Y, Zhou YJ, Tong GZ. Origin of highly pathogenic porcine reproductive and respiratory syndrome virus, China. Emerg Infect Dis. 2010; 16:365–367.
Article2. An TQ, Tian ZJ, Zhou YJ, Xiao Y, Peng JM, Chen J, Jiang YF, Hao XF, Tong GZ. Comparative genomic analysis of five pairs of virulent parental/attenuated vaccine strains of PRRSV. Vet Microbiol. 2011; 149:104–112.
Article3. An TQ, Zhou YJ, Liu GQ, Tian ZJ, Li J, Qiu HJ, Tong GZ. Genetic diversity and phylogenetic analysis of glycoprotein 5 of PRRSV isolates in mainland China from 1996 to 2006: coexistence of two NA-subgenotypes with great diversity. Vet Microbiol. 2007; 123:43–52.
Article4. Ansari IH, Kwon B, Osorio FA, Pattnaik AK. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J Virol. 2006; 80:3994–4004.
Article5. Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J Vet Diagn Invest. 1992; 4:127–133.
Article6. Cano JP, Dee SA, Murtaugh MP, Pijoan C. Impact of a modified-live porcine reproductive and respiratory syndrome virus vaccine intervention on a population of pigs infected with a heterologous isolate. Vaccine. 2007; 25:4382–4391.
Article7. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003; 31:3497–3500.
Article8. Costers S, Lefebvre DJ, Van Doorsselaere J, Vanhee M, Delputte PL, Nauwynck HJ. GP4 of porcine reproductive and respiratory syndrome virus contains a neutralizing epitope that is susceptible to immunoselection in vitro. Arch Virol. 2010; 155:371–378.
Article9. Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch Virol. 2000; 145:659–688.
Article10. Du Y, Lu Y, Qi J, Wu J, Wang G, Wang J. Complete genome sequence of a moderately pathogenic porcine reproductive and respiratory syndrome virus variant strain. J Virol. 2012; 86:13883–13884.
Article11. Faaberg KS, Even C, Palmer GA, Plagemann PG. Disulfide bonds between two envelope proteins of lactate dehydrogenase-elevating virus are essential for viral infectivity. J Virol. 1995; 69:613–617.
Article12. Fang Y, Kim DY, Ropp S, Steen P, Christopher-Hennings J, Nelson EA, Rowland RRR. Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. 2004; 100:229–235.
Article13. Han W, Wu JJ, Deng XY, Cao Z, Yu XL, Wang CB, Zhao TZ, Chen NH, Hu HH, Bin W, Hou LL, Wang LL, Tian KG, Zhang ZQ. Molecular mutations associated with the in vitro passage of virulent porcine reproductive and respiratory syndrome virus. Virus Genes. 2009; 38:276–284.
Article14. Holtkamp DJ, Kliebenstein JB, Neumann EJ, Zimmerman JJ, Rotto HF, Yoder TK, Wang C, Yeske PE, Mowrer CL, Haley CA. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J Swine Health Prod. 2013; 21:72–84.15. Huang C, Zhang Q, Feng WH. Regulation and evasion of antiviral immune responses by porcine reproductive and respiratory syndrome virus. Virus Res. 2015; 202:101–111.
Article16. Keffaber KK. Reproductive failure of unknown etiology. Am Assoc Swine Pract Newsl. 1989; 1:1–9.17. Leng CL, Tian ZJ, Zhang WC, Zhang HL, Zhai HY, An TQ, Peng JM, Ye C, Sun L, Wang Q, Sun Y, Li L, Zhao HY, Chang D, Cai XH, Zhang GH, Tong GZ. Characterization of two newly emerged isolates of porcine reproductive and respiratory syndrome virus from Northeast China in 2013. Vet Microbiol. 2014; 171:41–52.
Article18. Leng X, Li Z, Xia M, He Y, Wu H. Evaluation of the efficacy of an attenuated live vaccine against highly pathogenic porcine reproductive and respiratory syndrome virus in young pigs. Clin Vaccine Immunol. 2012; 19:1199–1206.
Article19. Li B, Fang L, Guo X, Gao J, Song T, Bi J, He K, Chen H, Xiao S. Epidemiology and evolutionary characteristics of the porcine reproductive and respiratory syndrome virus in China between 2006 and 2010. J Clin Microbiol. 2011; 49:3175–3183.
Article20. Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, Jiang W, Jiang P. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet J. 2007; 174:577–584.
Article21. Li Y, Wang X, Jiang P, Wang X, Chen W, Wang X, Wang K. Genetic variation analysis of porcine reproductive and respiratory syndrome virus isolated in China from 2002 to 2007 based on ORF5. Vet Microbiol. 2009; 138:150–155.
Article22. Li Y, Xue C, Wang L, Chen X, Chen F, Cao Y. Genomic analysis of two Chinese strains of porcine reproductive and respiratory syndrome viruses with different virulence. Virus Genes. 2010; 40:374–381.
Article23. Liu C, Ning Y, Xu B, Gong W, Zhang D. Analysis of genetic variation of porcine reproductive and respiratory syndrome virus (PRRSV) isolates in central China. J Vet Med Sci. 2016; 78:641–648.
Article24. Wenhui L, Zhongyan W, Guanqun Z, Zhili L, JingYun M, Qingmei X, Baoli S, Yingzuo B. Complete genome sequence of a novel variant porcine reproductive and respiratory syndrome virus (PRRSV) strain: evidence for recombination between vaccine and wild-type PRRSV strains. J Virol. 2012; 86:9543.
Article25. Meulenberg JJ, de Meijer EJ, Moormann RJ. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J Gen Virol. 1993; 74:1697–1701.
Article26. Meulenberg JJ, Hulst MM, de Meijer EJ, Moonen PL, den Besten A, de Kluyver EP, Wensvoort G, Moormann RJ. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993; 192:62–72.
Article27. Murtaugh MP, Xiao Z, Zuckermann F. Immunological responses of swine to porcine reproductive and respiratory syndrome virus infection. Viral Immunol. 2002; 15:533–547.
Article28. Nilubol D, Tripipat T, Hoonsuwan T, Tipsombatboon P, Piriyapongsa J. Genetic diversity of the ORF5 gene of porcine reproductive and respiratory syndrome virus (PRRSV) genotypes I and II in Thailand. Arch Virol. 2013; 158:943–953.
Article29. Plagemann PG. The primary GP5 neutralization epitope of North American isolates of porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol. 2004; 102:263–275.
Article30. Shi M, Lam TTY, Hon CC, Murtaugh MP, Davies PR, Hui RKH, Li J, Wong LTW, Yip CW, Jiang JW, Leung FCC. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J Virol. 2010; 84:8700–8711.
Article31. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739.
Article32. Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007; 2:e526.
Article33. Van Breedam W, Van Gorp H, Zhang JQ, Crocker PR, Delputte PL, Nauwynck HJ. The M/GP5 glycoprotein complex of porcine reproductive and respiratory syndrome virus binds the sialoadhesin receptor in a sialic acid-dependent manner. PLoS Pathog. 2010; 6:e1000730.
Article34. Wang FX, Qin LT, Liu Y, Liu X, Sun N, Yang Y, Chen T, Zhu HW, Ren JQ, Sun YJ, Cheng SP, Wen YJ. Novel Nsp2 deletion based on molecular epidemiology and evolution of porcine reproductive and respiratory syndrome virus in Shandong Province from 2013 to 2014. Infect Genet Evol. 2015; 33:219–226.
Article35. Wang X, He K, Zhang W, Zhou Z, Mao A, Yu Z. Genetic diversity and phylogenetic analysis of porcine reproductive and respiratory syndrome virus isolates in East China. Infect Genet Evol. 2014; 24:193–201.
Article36. Wei Z, Lin T, Sun L, Li Y, Wang X, Gao F, Liu R, Chen C, Tong G, Yuan S. N-linked glycosylation of GP5 of porcine reproductive and respiratory syndrome virus is critically important for virus replication in vivo. J Virol. 2012; 86:9941–9951.
Article37. Wensvoort G, Terpstra C, Pol JMA, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F, Broekhuijsen JM, Moonen PLJM, Zetstra T, de Boer EA, Tibben HJ, de Jong MF, van 't Veld P, Greenland GJR, van Gennep JA, Voets MTh, Verheijden JHM, Braamskamp J. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q. 1991; 13:121–130.
Article38. Wissink EH, van Wijk HA, Kroese MV, Weiland E, Meulenberg JJ, Rottier PJ, van Rijn PA. The major envelope protein, GP5, of a European porcine reproductive and respiratory syndrome virus contains a neutralization epitope in its N-terminal ectodomain. J Gen Virol. 2003; 84:1535–1543.
Article39. Xie J, Zhu W, Chen Y, Wei C, Zhou P, Zhang M, Huang Z, Sun L, Su S, Zhang G. Molecular epidemiology of PRRSV in South China from 2007 to 2011 based on the genetic analysis of ORF5. Microb Pathog. 2013; 63:30–36.
Article40. Yin G, Gao L, Shu X, Yang G, Guo S, Li W. Genetic diversity of the ORF5 gene of porcine reproductive and respiratory syndrome virus isolates in Southwest China from 2007 to 2009. PLoS One. 2012; 7:e33756.
Article41. Yuan S, Mickelson D, Murtaugh MP, Faaberg KS. Complete genome comparison of porcine reproductive and respiratory syndrome virus parental and attenuated strains. Virus Res. 2001; 79:189–200.
Article42. Yuan S, Nelsen CJ, Murtaugh MP, Schmitt BJ, Faaberg KS. Recombination between North American strains of porcine reproductive and respiratory syndrome virus. Virus Res. 1999; 61:87–98.
Article43. Zhou L, Chen S, Zhang J, Zeng J, Guo X, Ge X, Zhang D, Yang H. Molecular variation analysis of porcine reproductive and respiratory syndrome virus in China. Virus Res. 2009; 145:97–105.
Article44. Zhou L, Zhang J, Zeng J, Yin S, Li Y, Zheng L, Guo X, Ge X, Yang H. The 30-amino-acid deletion in the Nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J Virol. 2009; 83:5156–5167.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genetic diversity of porcine reproductive and respiratory syndrome virus in Korea
- Phylogenetic characterization of genes encoding for glycoprotein 5 and membrane protein of PRRSV isolate HH08
- A new recombined porcine reproductive and respiratory syndrome virus virulent strain in China
- Genetic diversity of nucleocapsid genes of recent porcine epidemic diarrhea viruses isolated in Korea
- Intracellular Localization of the Porcine Reproductive and Respiratory Syndrome Virus Nucleocapsid Protein