J Vet Sci.
2018 Jan;19(1):89-98. 10.4142/jvs.2018.19.1.89.
A new recombined porcine reproductive and respiratory syndrome virus virulent strain in China
- Affiliations
-
- 1Animal Science College & National Engineering Center for Swine Breeding Industry, South China Agriculture University, Guangzhou 510642, China. cxsong2004@163.com
- 2School of Animal Husbandry and Medical Engineering, Xinyang Agriculture and Forestry University, Xinyang 464000, China.
- KMID: 2402699
- DOI: http://doi.org/10.4142/jvs.2018.19.1.89
Abstract
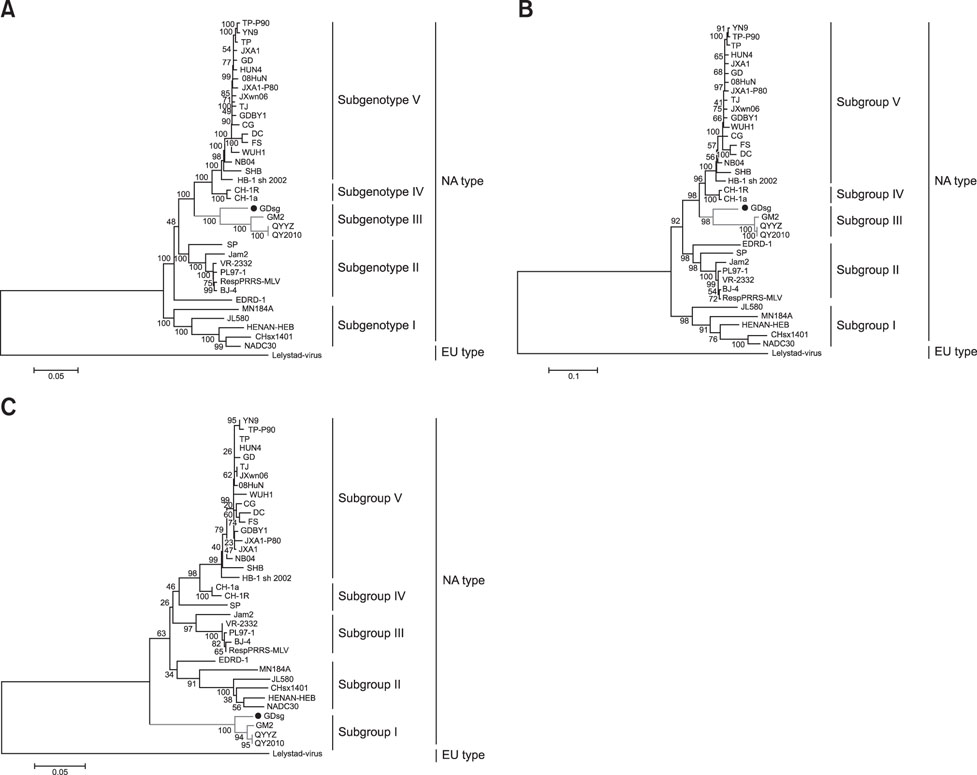
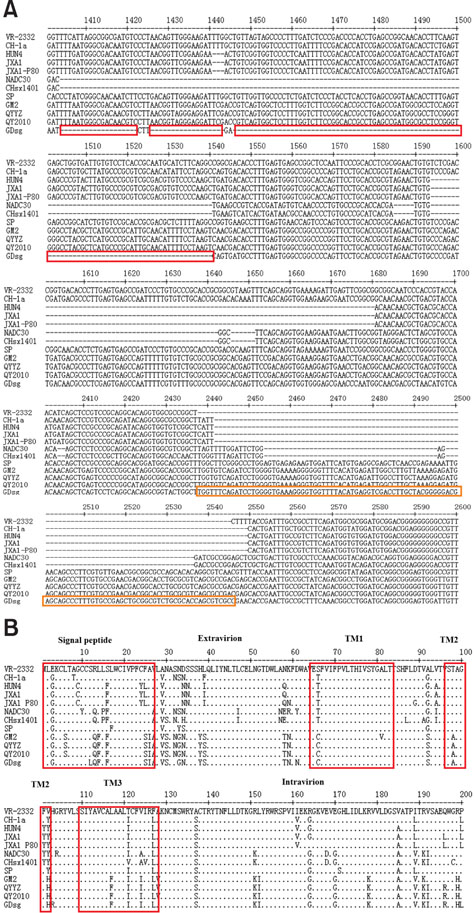
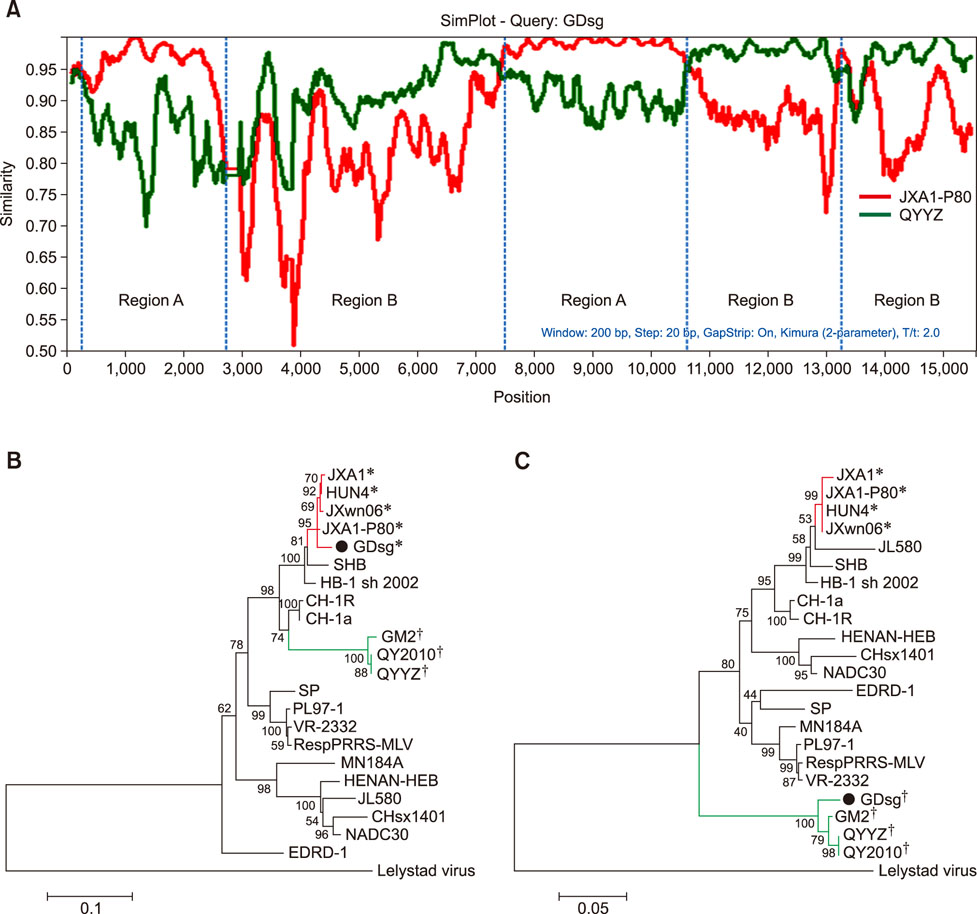
- Porcine reproductive and respiratory syndrome (PRRS) is one of the most important swine diseases worldwide. In the present study, a new virulent strain of PRRS virus (PRRSV), GDsg, was isolated in Guangdong province, China, and caused high fever, high morbidity, and high mortality in sows and piglets. The genome of this new strain was 15,413 nucleotides (nt) long, and comparative analysis revealed that GDsg shared 82.4% to 94% identity with type 2 PRRSV strains, but only 61.5% identity with type 1 PRRSV Lelystad virus strain. Phylogenetic analysis indicated that type 2 PRRSV isolates include five subgenotypes (I, II, III, IV, and V), which are represented by NADC30, VR-2332, GM2, CH-1a, and HuN4, respectively. Moreover, GDsg belongs to a newly emerging type 2 PRRSV subgenotype III. More interestingly, the newly isolated GDsg strain has multiple discontinuous nt deletions, 131 (19 + 18 + 94) at position 1404-1540 and a 107 nt insertion in the NSP2 region. Most importantly, the GDsg strain was identified as a virus recombined between low pathogenic field strain QYYZ and vaccine strain JXA1-P80. In conclusion, a new independent subgenotype and recombinant PRRSV strain has emerged in China and could be a new threat to the swine industry of China.
Keyword
MeSH Terms
Figure
Reference
-
1. Albina E. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet Microbiol. 1997; 55:309–316.
Article2. Ansari IH, Kwon B, Osorio FA, Pattnaik AK. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J Virol. 2006; 80:3994–4004.
Article3. Bautista EM, Faaberg KS, Mickelson D, McGruder ED. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology. 2002; 298:258–270.
Article4. Bautista EM, Meulenberg JJ, Choi CS, Molitor TW. Structural polypeptides of the American (VR-2332) strain of porcine reproductive and respiratory syndrome virus. Arch Virol. 1996; 141:1357–1365.
Article5. Beura LK, Sarkar SN, Kwon B, Subramaniam S, Jones C, Pattnaik AK, Osorio FA. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J Virol. 2010; 84:1574–1584.
Article6. Chang CC, Yoon KJ, Zimmerman JJ, Harmon KM, Dixon PM, Dvorak CM, Murtaugh MP. Evolution of porcine reproductive and respiratory syndrome virus during sequential passages in pigs. J Virol. 2002; 76:4750–4763.
Article7. Conzelmann KK, Visser N, Van Woensel P, Thiel HJ. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology. 1993; 193:329–339.
Article8. Deng Y, Pan Y, Wang D, Zhou Q, Bi Y, Chen F, Song Y. Complete genome sequence of porcine reproductive and respiratory syndrome virus strain QY2010 reveals a novel subgroup emerging in China. J Virol. 2012; 86:7719–7720.
Article9. Feng Y, Zhao T, Nguyen T, Inui K, Ma Y, Nguyen TH, Nguyen VC, Liu D, Bui QA, To LT, Wang C, Tian K, Gao GF. Porcine respiratory and reproductive syndrome virus variants, Vietnam and China, 2007. Emerg Infect Dis. 2008; 14:1774–1776.
Article10. Forsberg R. Divergence time of porcine reproductive and respiratory syndrome virus subtypes. Mol Biol Evol. 2005; 22:2131–2134.
Article11. Han J, Wang Y, Faaberg KS. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 2006; 122:175–182.
Article12. Ji G, Li Y, Tan F, Zhuang J, Li X, Tian K. Complete genome sequence of an NADC30-like strain of porcine reproductive and respiratory syndrome virus in China. Genome Announc. 2016; 4:e00303–e00316.
Article13. Kapur V, Elam MR, Pawlovich TM, Murtaugh MP. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the midwestern United States. J Gen Virol. 1996; 77:1271–1276.
Article14. Li B, Fang L, Guo X, Gao J, Song T, Bi J, He K, Chen H, Xiao S. Epidemiology and evolutionary characteristics of the porcine reproductive and respiratory syndrome virus in China between 2006 and 2010. J Clin Microbiol. 2011; 49:3175–3183.
Article15. Li B, Fang L, Xu Z, Liu S, Gao J, Jiang Y, Chen H, Xiao S. Recombination in vaccine and circulating strains of porcine reproductive and respiratory syndrome viruses. Emerg Infect Dis. 2009; 15:2032–2035.
Article16. Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, Jiang W, Jiang P. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet J. 2007; 174:577–584.
Article17. Li Y, Zhou L, Zhang J, Ge X, Zhou R, Zheng H, Geng G, Guo X, Yang H. Nsp9 and Nsp10 contribute to the fatal virulence of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China. PLoS Pathog. 2014; 10:e1004216.
Article18. Liu D, Zhou R, Zhang J, Zhou L, Jiang Q, Guo X, Ge X, Yang H. Recombination analyses between two strains of porcine reproductive and respiratory syndrome virus in vivo. Virus Res. 2011; 155:473–486.
Article19. Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999; 73:152–160.
Article20. Lu WH, Tun HM, Sun BL, Mo J, Zhou QF, Deng YX, Xie QM, Bi YZ, Leung FC, Ma JY. Re-emerging of porcine respiratory and reproductive syndrome virus (lineage 3) and increased pathogenicity after genomic recombination with vaccine variant. Vet Microbiol. 2015; 175:332–340.
Article21. Meulenberg JJ, Hulst MM, de Meijer EJ, Moonen PL, den Besten A, de Kluyver EP, Wensvoort G, Moormann RJ. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993; 192:62–72.
Article22. Murtaugh MP, Elam MR, Kakach LT. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch Virol. 1995; 140:1451–1460.
Article23. Nelsen CJ, Murtaugh MP, Faaberg KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. 1999; 73:270–280.
Article24. Nelson EA, Christopher-Hennings J, Drew T, Wensvoort G, Collins JE, Benfield DA. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J Clin Microbiol. 1993; 31:3184–3189.
Article25. Pejsak Z, Stadejek T, Markowska-Daniel I. Clinical signs and economic losses caused by porcine reproductive and respiratory syndrome virus in a large breeding farm. Vet Microbiol. 1997; 55:317–322.
Article26. Shi M, Holmes EC, Brar MS, Leung FC. Recombination is associated with an outbreak of novel highly pathogenic porcine reproductive and respiratory syndrome viruses in China. J Virol. 2013; 87:10904–10907.
Article27. Song C, Krell P, Yoo D. Nonstructural protein 1α subunit-based inhibition of NF-κB activation and suppression of interferon-β production by porcine reproductive and respiratory syndrome virus. Virology. 2010; 407:268–280.
Article28. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739.
Article29. Wenhui L, Zhongyan W, Guanqun Z, Zhili L, JingYun M, Qingmei X, Baoli S, Yingzuo B. Complete genome sequence of a novel variant porcine reproductive and respiratory syndrome virus (PRRSV) strain: evidence for recombination between vaccine and wild-type PRRSV strains. J Virol. 2012; 86:9543.
Article30. Wesley RD, Mengeling WL, Lager KM, Vorwald AC, Roof MB. Evidence for divergence of restriction fragment length polymorphism patterns following in vivo replication of porcine reproductive and respiratory syndrome virus. Am J Vet Res. 1999; 60:463–467.31. Xie J, Zhu W, Chen Y, Wei C, Zhou P, Zhang M, Huang Z, Sun L, Su S, Zhang G. Molecular epidemiology of PRRSV in South China from 2007 to 2011 based on the genetic analysis of ORF5. Microb Pathog. 2013; 63:30–36.
Article32. Yuan S, Nelsen CJ, Murtaugh MP, Schmitt BJ, Faaberg KS. Recombination between North American strains of porcine reproductive and respiratory syndrome virus. Virus Res. 1999; 61:87–98.
Article33. Zhao K, Ye C, Chang XB, Jiang CG, Wang SJ, Cai XH, Tong GZ, Tian ZJ, Shi M, An TQ. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J Virol. 2015; 89:10712–10716.
Article34. Zhou L, Chen S, Zhang J, Zeng J, Guo X, Ge X, Zhang D, Yang H. Molecular variation analysis of porcine reproductive and respiratory syndrome virus in China. Virus Res. 2009; 145:97–105.
Article35. Zhou L, Wang Z, Ding Y, Ge X, Guo X, Yang H. NADC30-like strain of porcine reproductive and respiratory syndrome virus, China. Emerg Infect Dis. 2015; 21:2256–2257.
Article36. Zhou L, Yang H. Porcine reproductive and respiratory syndrome in China. Virus Res. 2010; 154:31–37.
Article37. Zhou L, Yang X, Tian Y, Yin S, Geng G, Ge X, Guo X, Yang H. Genetic diversity analysis of genotype 2 porcine reproductive and respiratory syndrome viruses emerging in recent years in China. Biomed Res Int. 2014; 2014:748068.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Evaluation of the efficacy of an attenuated live vaccine based on virulent porcine reproductive and respiratory syndrome virus 2 in young pigs
- Intracellular Localization of the Porcine Reproductive and Respiratory Syndrome Virus Nucleocapsid Protein
- Genetic diversity and phylogenetic analysis of porcine reproductive and respiratory syndrome virus in southern China from 2007 to 2014
- Genomic characterization and pathogenic study of two porcine reproductive and respiratory syndrome viruses with different virulence in Fujian, China
- A survey of porcine reproductive and respiratory syndrome among wild boar populations in Korea