Immune Netw.
2017 Dec;17(6):410-423. 10.4110/in.2017.17.6.410.
Terminally Differentiating Eosinophils Express Neutrophil Primary Granule Proteins as well as Eosinophil-specific Granule Proteins in a Temporal Manner
- Affiliations
-
- 1Department of Bionano Engineering, Hanyang University, Ansan 15588, Korea. iychu@hanyang.ac.kr
- 2Division of Allergy and Respiratory Medicine, Department of Internal Medicine, Soonchunhyang University Bucheon Hospital, Bucheon 14584, Korea.
- 3Department of Microbiology, Gachon University School of Medicine, Incheon 21936, Korea.
- 4Department of Molecular and Life Sciences, Hanyang University, Ansan 15588, Korea.
- KMID: 2400627
- DOI: http://doi.org/10.4110/in.2017.17.6.410
Abstract
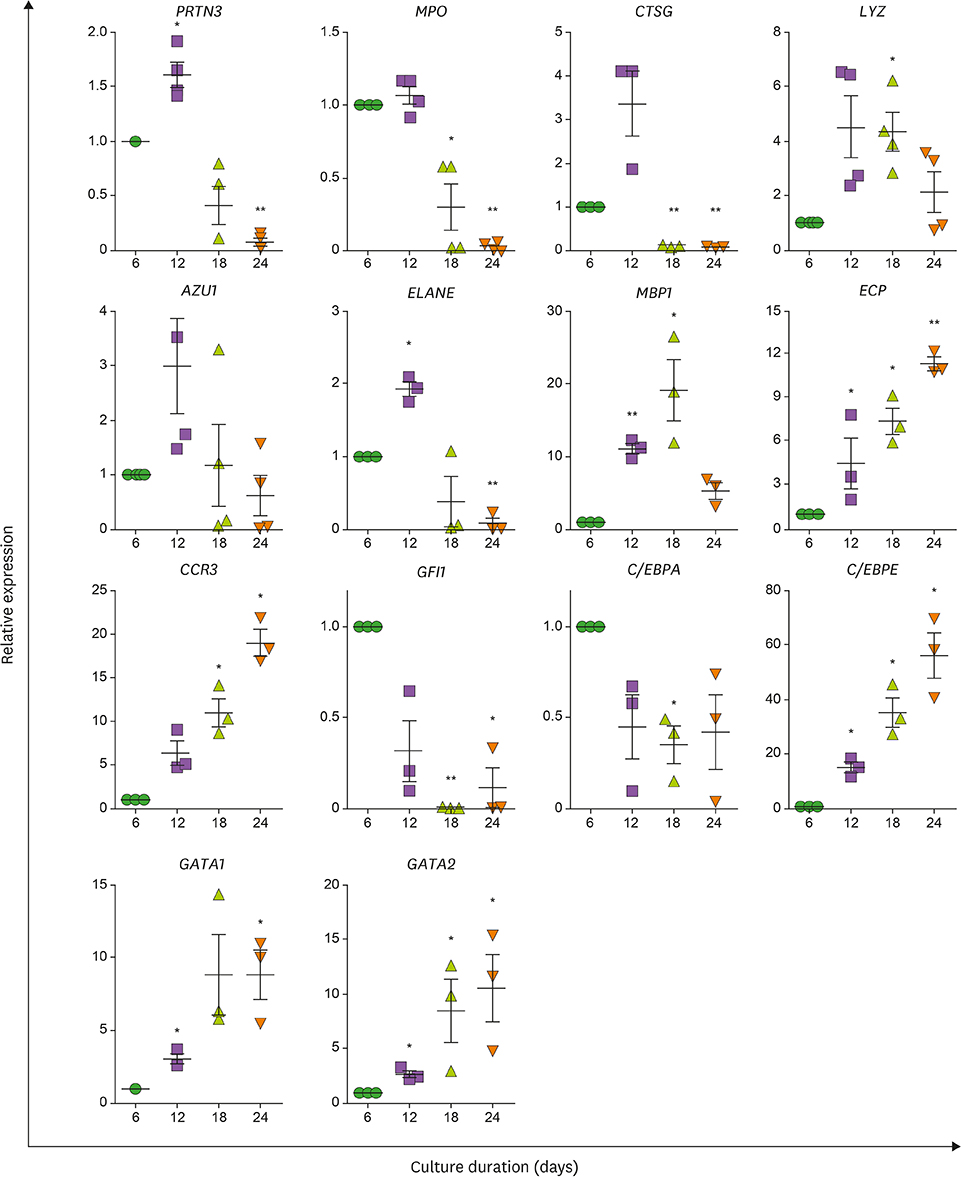
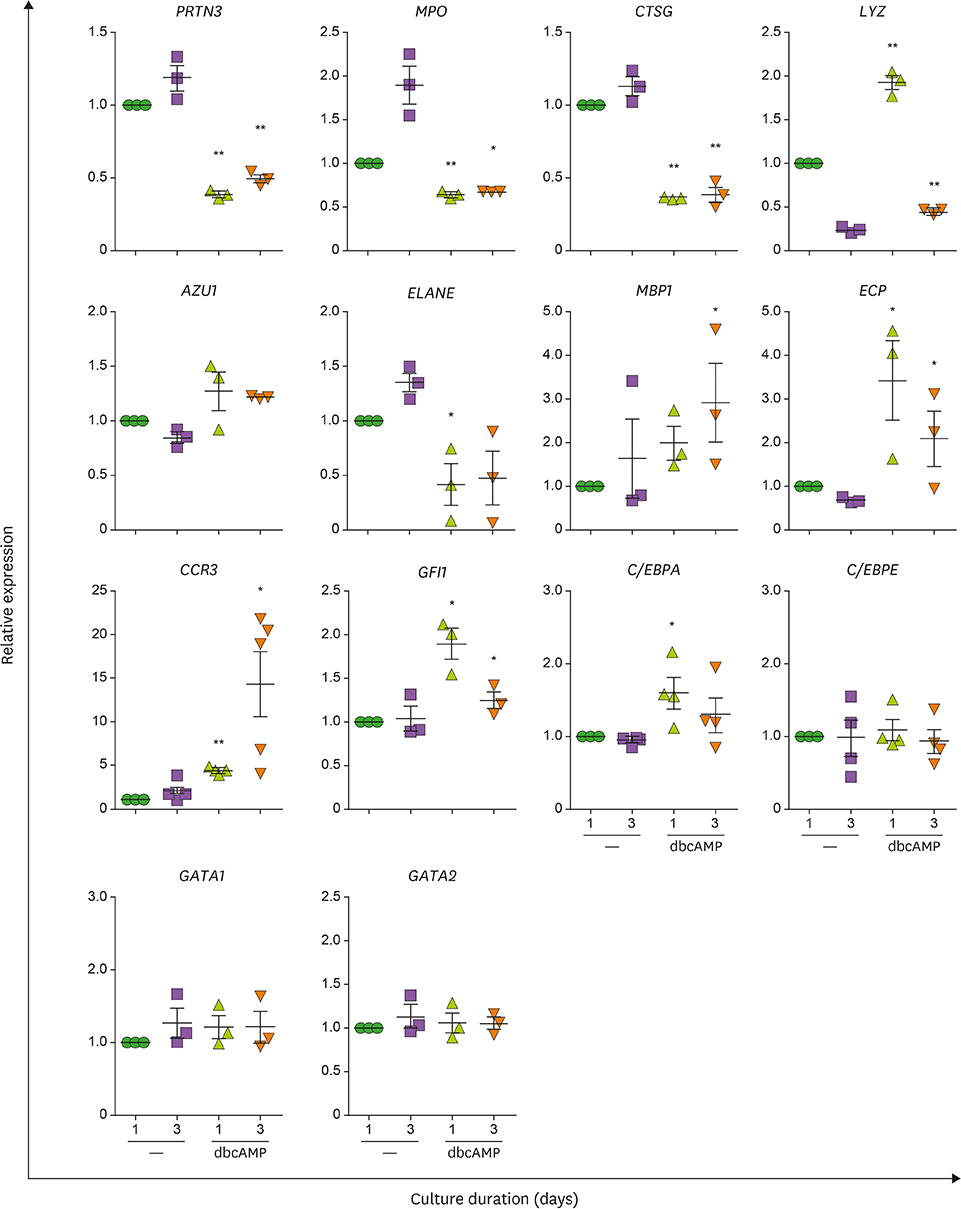
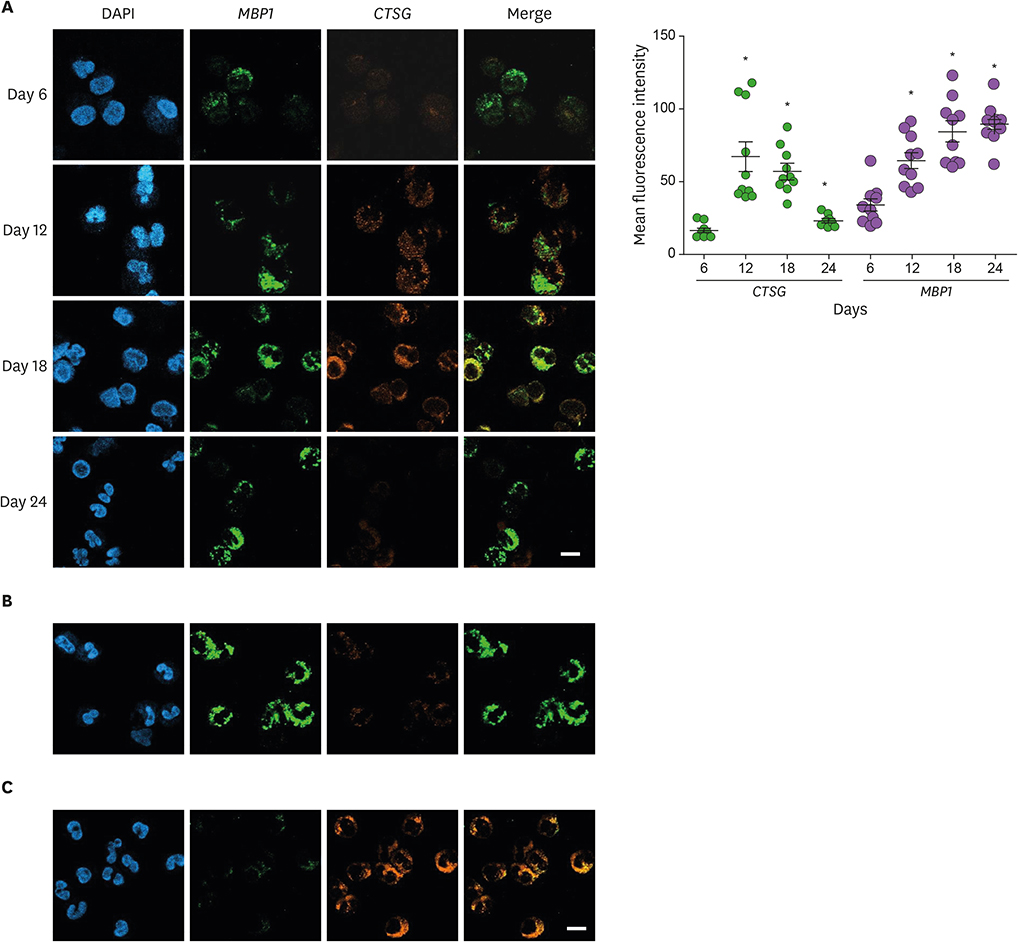
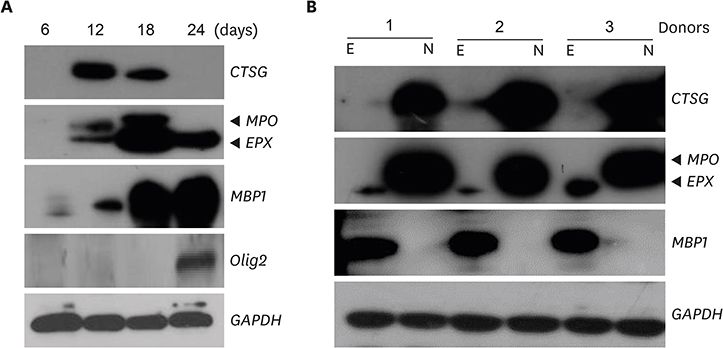
- Neutrophils and eosinophils, 2 prominent granulocytes, are commonly derived from myelocytic progenitors through successive stages in the bone marrow. Our previous genome-wide transcriptomic data unexpectedly showed that genes encoding a multitude of neutrophil primary granule proteins (NPGPs) were markedly downregulated during the end period of eosinophilic terminal differentiation when cord blood (CB) cluster of differentiation (CD) 34+ cells were induced to differentiate toward the eosinophil lineage during a 24-day culture period. Accordingly, this study aimed to examine whether NPGP genes were expressed on the way to eosinophil terminal differentiation stage and to compare their expression kinetics with that of genes encoding eosinophil-specific granule proteins (ESGPs). Transcripts of all NPGP genes examined, including proteinase 3, myeloperoxidase, cathepsin G (CTSG), and neutrophil elastase, reached a peak at day 12 and sharply declined thereafter, while transcript of ESGP genes including major basic protein 1 (MBP1) attained maximum expression at days 18 or 24. Growth factor independent 1 (GFI1) and CCAAT/enhancer-binding protein α (C/EBPA), transactivators for the NPGP genes, were expressed immediately before the NPGP genes, whereas expression of C/EBPA, GATA1, and GATA2 kinetically paralleled that of eosinophil granule protein genes. The expression kinetics of NPGPs and ESGPs were duplicated upon differentiation of the eosinophilic leukemia cell line (EoL-1) immature eosinophilic cells. Importantly, confocal image analysis showed that CTSG was strongly coexpressed with MBP1 in differentiating CB eosinophils at days 12 and 18 and became barely detectable at day 24 and beyond. Our results suggest for the first time the presence of an immature stage where eosinophils coexpress NPGPs and ESGPs before final maturation.
Keyword
MeSH Terms
Figure
Reference
-
1. Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 2012; 30:459–489.
Article2. Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006; 24:147–174.
Article3. Willebrand R, Voehringer D. Regulation of eosinophil development and survival. Curr Opin Hematol. 2017; 24:9–15.
Article4. Fiedler K, Brunner C. The role of transcription factors in the guidance of granulopoiesis. Am J Blood Res. 2012; 2:57–65.5. Mori Y, Iwasaki H, Kohno K, Yoshimoto G, Kikushige Y, Okeda A, Uike N, Niiro H, Takenaka K, Nagafuji K, et al. Identification of the human eosinophil lineage-committed progenitor: revision of phenotypic definition of the human common myeloid progenitor. J Exp Med. 2009; 206:183–193.
Article6. Borregaard N, Cowland JB. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood. 1997; 89:3503–3521.
Article7. Borregaard N, Theilgaard-Mönch K, Sørensen OE, Cowland JB. Regulation of human neutrophil granule protein expression. Curr Opin Hematol. 2001; 8:23–27.
Article8. Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992; 80:1725–1735.
Article9. D’Alo’ F, Johansen LM, Nelson EA, Radomska HS, Evans EK, Zhang P, Nerlov C, Tenen DG. The amino terminal and E2F interaction domains are critical for C/EBP alpha-mediated induction of granulopoietic development of hematopoietic cells. Blood. 2003; 102:3163–3171.10. Gombart AF, Kwok SH, Anderson KL, Yamaguchi Y, Torbett BE, Koeffler HP. Regulation of neutrophil and eosinophil secondary granule gene expression by transcription factors C/EBP epsilon and PU.1. Blood. 2003; 101:3265–3273.
Article11. Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010; 33:657–670.
Article12. Theilgaard-Mönch K, Jacobsen LC, Borup R, Rasmussen T, Bjerregaard MD, Nielsen FC, Cowland JB, Borregaard N. The transcriptional program of terminal granulocytic differentiation. Blood. 2005; 105:1785–1796.
Article13. Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, Orkin SH. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003; 18:109–120.
Article14. Dvorak AM, Letourneau L, Login GR, Weller PF, Ackerman SJ. Ultrastructural localization of the Charcot-Leyden crystal protein (lysophospholipase) to a distinct crystalloid-free granule population in mature human eosinophils. Blood. 1988; 72:150–158.
Article15. Du J, Stankiewicz MJ, Liu Y, Xi Q, Schmitz JE, Lekstrom-Himes JA, Ackerman SJ. Novel combinatorial interactions of GATA-1, PU.1, and C/EBPepsilon isoforms regulate transcription of the gene encoding eosinophil granule major basic protein. J Biol Chem. 2002; 277:43481–43494.
Article16. Uhm TG, Kim BS, Chung IY. Eosinophil development, regulation of eosinophil-specific genes, and role of eosinophils in the pathogenesis of asthma. Allergy Asthma Immunol Res. 2012; 4:68–79.
Article17. Hwang SM, Uhm TG, Lee SK, Kong SK, Jung KH, Binas B, Chai YG, Park SW, Chung IY. Olig2 is expressed late in human eosinophil development and controls Siglec-8 expression. J Leukoc Biol. 2016; 100:711–723.
Article18. Uhm TG, Lee SK, Kim BS, Kang JH, Park CS, Rhim TY, Chang HS, Kim DJ, Chung IY. CpG methylation at GATA elements in the regulatory region of CCR3 positively correlates with CCR3 transcription. Exp Mol Med. 2012; 44:268–280.
Article19. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012; 9:671–675.
Article20. Lee JJ, Jacobsen EA, Ochkur SI, McGarry MP, Condjella RM, Doyle AD, Luo H, Zellner KR, Protheroe CA, Willetts L, et al. Human versus mouse eosinophils: “that which we call an eosinophil, by any other name would stain as red. J Allergy Clin Immunol. 2012; 130:572–584.
Article21. Pillay J, Tak T, Kamp VM, Koenderman L. Immune suppression by neutrophils and granulocytic myeloid-derived suppressor cells: similarities and differences. Cell Mol Life Sci. 2013; 70:3813–3827.
Article22. Levi-Schaffer F, Lacy P, Severs NJ, Newman TM, North J, Gomperts B, Kay AB, Moqbel R. Association of granulocyte-macrophage colony-stimulating factor with the crystalloid granules of human eosinophils. Blood. 1995; 85:2579–2586.
Article23. Nakajima T, Matsumoto K, Suto H, Tanaka K, Ebisawa M, Tomita H, Yuki K, Katsunuma T, Akasawa A, Hashida R, et al. Gene expression screening of human mast cells and eosinophils using high-density oligonucleotide probe arrays: abundant expression of major basic protein in mast cells. Blood. 2001; 98:1127–1134.
Article24. Mayumi M. EoL-1, a human eosinophilic cell line. Leuk Lymphoma. 1992; 7:243–250.
Article25. Wong CK, Ho CY, Lam CW, Zhang JP, Hjelm NM. Differentiation of a human eosinophilic leukemic cell line, EoL-1: characterization by the expression of cytokine receptors, adhesion molecules, CD95 and eosinophilic cationic protein (ECP). Immunol Lett. 1999; 68:317–323.
Article26. Jung Y. Comparative analysis of dibutyric cAMP and butyric acid on the differentiation of human eosinophilic leukemia EoL-1 cells. Immune Netw. 2015; 15:313–318.
Article27. Sakamaki K, Kanda N, Ueda T, Aikawa E, Nagata S. The eosinophil peroxidase gene forms a cluster with the genes for myeloperoxidase and lactoperoxidase on human chromosome 17. Cytogenet Cell Genet. 2000; 88:246–248.
Article28. Sakamaki K, Tomonaga M, Tsukui K, Nagata S. Molecular cloning and characterization of a chromosomal gene for human eosinophil peroxidase. J Biol Chem. 1989; 264:16828–16836.
Article29. Wilkerson EM, Johansson MW, Hebert AS, Westphall MS, Mathur SK, Jarjour NN, Schwantes EA, Mosher DF, Coon JJ. The peripheral blood eosinophil proteome. J Proteome Res. 2016; 15:1524–1533.
Article30. Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren-Shaul H, Winter D, Lara-Astiaso D, Gury M, Weiner A, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015; 163:1663–1677.
Article31. Wong TW, Jelinek DF. Purification of functional eosinophils from human bone marrow. J Immunol Methods. 2013; 387:130–139.
Article32. Dyer KD, Moser JM, Czapiga M, Siegel SJ, Percopo CM, Rosenberg HF. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J Immunol. 2008; 181:4004–4009.
Article33. Lominadze G, Powell DW, Luerman GC, Link AJ, Ward RA, McLeish KR. Proteomic analysis of human neutrophil granules. Mol Cell Proteomics. 2005; 4:1503–1521.
Article34. Liu Q, Dong F. Gfi-1 inhibits the expression of eosinophil major basic protein (MBP) during G-CSF-induced neutrophilic differentiation. Int J Hematol. 2012; 95:640–647.
Article35. El Ouriaghli F, Fujiwara H, Melenhorst JJ, Sconocchia G, Hensel N, Barrett AJ. Neutrophil elastase enzymatically antagonizes the in vitro action of G-CSF: implications for the regulation of granulopoiesis. Blood. 2003; 101:1752–1758.
Article36. Avalos BR. Molecular analysis of the granulocyte colony-stimulating factor receptor. Blood. 1996; 88:761–777.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Emerging Role of Eosinophils as Multifunctional Leukocytes in Health and Disease
- Toluene Diisocyanate-Induced Allergic Rhinitis in Guinea Pigs: Investigating the Relation between Eosinophil Infiltration and Epithelial Injury
- Serum Eosinophil Cationic Protein Levels in Patients with Allergic Diseases
- The Electron Microscopic Study of Enzymes in Eosinophils
- Utility of eosinophil granule proteins in management of pediatric chronic cough