J Vet Sci.
2012 Dec;13(4):405-412.
Molecular characterization of two Bangladeshi infectious bursal disease virus isolates using the hypervariable sequence of VP2 as a genetic marker
- Affiliations
-
- 1Department of Microbiology and Hygiene, Faculty of Veterinary Science, Bangladesh Agricultural University, Mymensingh 2202, Bangladesh. islammtuq@yahoo.com
- 2Department of Medicine, Faculty of Veterinary Science, Bangladesh Agricultural University, Mymensingh 2202, Bangladesh.
- 3Department of Immunology, Institute of Biotechnology, Hanoi 10000, Vietnam.
- 4Department of Medical Microbiology and Immunology, Faculty of Medicine, National University of Malaysia, Kuala Lumpur 43600, Malaysia.
- 5School of Sustainable Agriculture, University Malaysia Sabah, Kota Kinabalu 88400, Malaysia.
Abstract
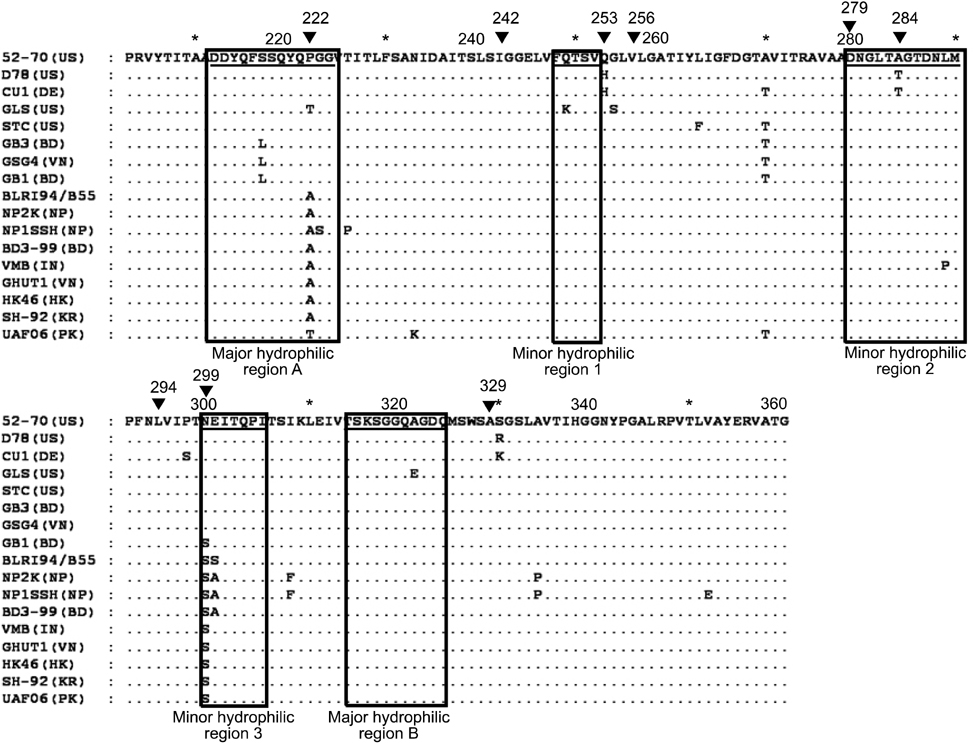
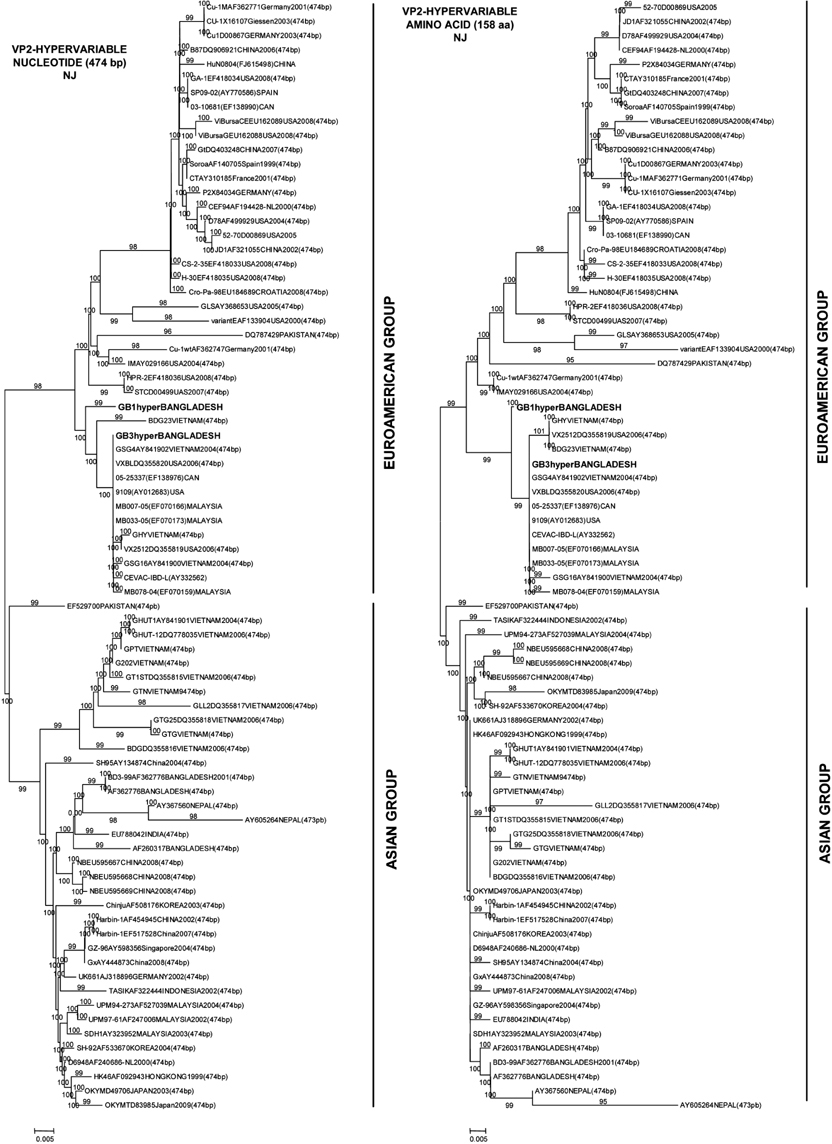
- Two Bangladeshi infectious bursal disease virus (IBDV) isolates collected in 2007, termed GB1 and GB3, were subjected to comparative sequencing and phylogenetic analyses. Sequence analysis of a 474-bp hypervariable region in the VP2 gene revealed that among four major amino acid substitutions observed in the strains, two were unique to GB1 and GB3 (Ser217Leu and Ala270Thr) while one substitution was only found in GB1 (Asn299Ser). Among IBDVs from Bangladesh including GB1 and GB3, the rate of identity and homology was around 97~99%. The amino acid sequences of GB1 and GB3 differ from those of previous Bangladeshi IBDV isolates and contain amino acid substitutions Pro222Ala and Asn299Ser (in GB3 only). Phylogenetic analysis revealed that GB1 and GB3 are grouped with other very virulent IBDVs of European and American origin in contrast to two previously isolated Bangladeshi IBDV strains (GenBank accession Nos. AF362776 and AF260317), which belong to the Asian group. It was concluded that GB1 and GB3 belong to a very virulent group of IBDVs. However, amino acid sequences of GB1 and GB3 differ from those of the other Bangladeshi IBDVs by one or two amino acids encoded in the hypervariable region of the VP2 gene.
MeSH Terms
Figure
Reference
-
1. Biswas PK, Biswas D, Ahmed S, Rahman A, Debnath NC. A longitudinal study of the incidence of major endemic and epidemic diseases affecting semi-scavenging chickens reared under the Participatory Livestock Development Project areas in Bangladesh. Avian Pathol. 2005. 34:303–312.
Article2. Boot HJ, Hoekman AJW, Gielkens ALJ. The enhanced virulence of very virulent infectious bursal disease virus is partly determined by its B-segment. Arch Virol. 2005. 150:137–144.
Article3. Boot HJ, ter Huurne AAHM, Hoekman AJW, Peeters BPH, Gielkens ALJ. Rescue of very virulent and mosaic infectious bursal disease virus from cloned cDNA: VP2 is not the sole determinant of the very virulent phenotype. J Virol. 2000. 74:6701–6711.
Article4. Cosgrove AS. An apparently new disease of chickens - avian nephrosis. Avian Dis. 1962. 6:385–389.5. Eterradossi N, Arnauld C, Toquin D, Rivallan G. Critical amino acid changes in VP2 variable domain are associated with typical and atypical antigenicity in very virulent infectious bursal disease viruses. Arch Virol. 1998. 143:1627–1636.
Article6. Ignjatovic J, Sapats S. Confirmation of the existence of two distinct genetic groups of infectious bursal disease virus in Australia. Aust Vet J. 2002. 80:689–694.
Article7. Islam MR, Chowdhury EH, Das PM, Dewan ML. Pathology of acute infectious bursal disease virus in chickens induced experimentally with a very virulent isolate. Indian J Anim Sci. 1997. 67:7–9.8. Islam MR, Zierenberg K, Eterradossi N, Toquin D, Rivallan G, Müller H. Molecular and antigenic characterization of Bangladeshi isolates of infectious bursal disease virus demonstrate their similarities with recent European, Asian and African very virulent strains. J Vet Med B Infect Dis Vet Public Health. 2001. 48:211–221.
Article9. Jackwood DJ. Recent trends in the molecular diagnosis of infectious bursal disease viruses. Anim Health Res Rev. 2004. 5:313–316.
Article10. Jackwood DJ, Sreedevi B, LeFever LJ, Sommer-Wagner SE. Studies on naturally occurring infectious bursal disease viruses suggest that a single amino acid substitution at position 253 in VP2 increases pathogenicity. Virology. 2008. 377:110–116.
Article11. Kibenge FSB, Jackwood DJ, Mercado CC. Nucleotide sequence analysis of genome segment A of infectious bursal disease virus. J Gen Virol. 1990. 71:Pt 3. 569–577.
Article12. Kibenge FSB, Qian B, Cleghorn JR, Martin CK. Infectious bursal disease virus polyprotein processing does not involve cellular proteases. Arch Virol. 1997. 142:2401–2419.
Article13. Leong JC, Brown D, Dobos P, Kibenge FSB, Ludert JE, Müller H, Mundt E, Nicholson B. van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB, editors. Familly Birnaviridae. Virus Taxonomy: Classification and Nomenclature of Viruses: Seventh Report of the International Committee on Taxonomy of Viruses. 2000. San Diego: Academic Press;481–490.14. Liu M, Vakharia VN. VP1 protein of infectious bursal disease virus modulates the virulence in vivo. Virology. 2004. 330:62–73.
Article15. Lukert PD, Saif YM. Saif YM, Barnes HJ, Glisson JR, Fadly AM, McDougald LR, Swayne DE, editors. Infectious bursal disease. Diseases of Poultry. 2003. 11th ed. Ames: Iowa State Press;161–179.16. McFerran JB, McNulty MS, McKillop ER, Connor TJ, McCracken RM, Collins DS, Allan GM. Isolation and serological studies with infectious bursal disease viruses from fowl, turkeys and ducks: demonstration of a second serotype. Avian Pathol. 1980. 9:395–404.
Article17. Mundt E, Köllner B, Kretzschmar D. VP5 of infectious bursal disease virus is not essential for viral replication in cell culture. J Virol. 1997. 71:5647–5651.
Article18. Müller H, Islam MR, Raue R. Research on infectious bursal disease--the past, the present and the future. Vet Microbiol. 2003. 97:153–165.
Article19. Müller R, Käufer I, Reinacher M, Weiss E. Immunofluorescent studies of early virus propagation after oral infection with infectious bursal disease virus (IBDV). Zentralbl Veterinarmed B. 1979. 26:345–352.
Article20. Nagarajan MM, Kibenge FSB. Infectious bursal disease virus: a review of molecular basis for variations in antigenicity and virulence. Can J Vet Res. 1997. 61:81–88.21. Nicholas KB, Nicholas HB Jr, Deerfield DW 2nd. GeneDoc: analysis and visualization of genetic variation. EMBNET News. 1997. 4:1–4.22. Qi X, Gao H, Gao Y, Qin L, Wang Y, Gao L, Wang X. Naturally occurring mutations at residues 253 and 284 in VP2 contribute to the cell tropism and virulence of very virulent infectious bursal disease virus. Antiviral Res. 2009. 84:225–233.
Article23. Rautenschlein S, Yeh HY, Sharma JM. Comparative immunopathogenesis of mild, intermediate, and virulent strains of classic infectious bursal disease virus. Avian Dis. 2003. 47:66–78.
Article24. Rodriguez-Chavez IR, Rosenberger JK, Cloud SS. Characterization of the antigenic, immunogenic, and pathogenic variation of infectious bursal disease virus due to propagation in different host systems (bursa, embryo, and cell culture). II. Antigenicity at the epitope level. Avian Pathol. 2002. 31:473–483.
Article25. Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. 2001. Vol. 3:3rd ed. New York: Cold Spring Harbor Laboratory Press;15.1–18.136.26. Sánchez AB, Rodriguez JF. Proteolytic processing in infectious bursal disease virus: identification of the polyprotein cleavage sites by site-directed mutagenesis. Virology. 1999. 262:190–199.
Article27. Schnitzler D, Bernstein F, Müller H, Becht H. The genetic basis for the antigenicity of the VP2 protein of the infectious bursal disease virus. J Gen Virol. 1993. 74(Pt 8):1563–1571.
Article28. Spies U, Müller H. Demonstration of enzyme activities required for cap structure formation in infectious bursal disease virus, a member of the birnavirus group. J Gen Virol. 1990. 71(Pt 4):977–981.
Article29. Tacken MGJ, Peeters BPH, Thomas AAM, Rottier PJM, Boot HJ. Infectious bursal disease virus capsid protein VP3 interacts both with VP1, the RNA-dependent RNA polymerase, and with viral double-stranded RNA. J Virol. 2002. 76:11301–11311.
Article30. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007. 24:1596–1599.
Article31. van den Berg TP, Gonze M, Meulemans G. Acute infectious bursal disease in poultry: isolation and characterisation of a highly virulent strain. Avian Pathol. 1991. 20:133–143.
Article32. van den Berg TP, Gonze M, Morales D, Meulemans G. Acute infectious bursal disease in poultry: immunological and molecular basis of antigenicity of a highly virulent strain. Avian Pathol. 1996. 25:751–768.
Article33. van den Berg TP, Morales D, Eterradossi N, Rivallan G, Toquin D, Raue R, Zierenberg K, Zhang MF, Zhu YP, Wang CQ, Zheng HJ, Wang X, Chen GC, Lim BL, Müller H. Assessment of genetic, antigenic and pathotypic criteria for the characterization of IBDV strains. Avian Pathol. 2004. 33:470–476.
Article34. Wang XM, Zeng XW, Gao HL, Fu CY, Wei P. Changes in VP2 gene during the attenuation of very virulent infectious bursal disease virus strain Gx isolated in China. Avian Dis. 2004. 48:77–83.
Article35. Yamaguchi T, Kasanga CJ, Terasaki K, Maw MT, Ohya K, Fukushi H. Nucleotide sequence analysis of VP2 hypervariable domain of infectious bursal disease virus detected in Japan from 1993 to 2004. J Vet Med Sci. 2007. 69:733–738.
Article36. Yamaguchi T, Ogawa M, Inoshima Y, Miyoshi M, Fukushi H, Hirai K. Identification of sequence changes responsible for the attenuation of highly virulent infectious bursal disease virus. Virology. 1996. 223:219–223.
Article37. Yamaguchi T, Ogawa M, Miyoshi M, Inoshima Y, Fukushi H, Hirai K. Sequence and phylogenetic analyses of highly virulent infectious bursal disease virus. Arch Virol. 1997. 142:1441–1458.
Article38. Zierenberg K, Nieper H, van den Berg TP, Ezeokoli CD, Voss M, Müller H. The VP2 variable region of African and German isolates of infectious bursal disease virus: comparison with very virulent, "classical" virulent, and attenuated tissue culture-adapted strains. Arch Virol. 2000. 145:113–125.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Efficacy of VP2 protein expressed in E. coli for protection against highly virulent infectious bursal disease virus
- Evaluation of modified vaccinia virus Ankara expressing VP2 protein of infectious bursal disease virus as an immunogen in chickens
- Sequence analysis of segment A gene of a very virulent infectious bursal disease virus recently isolated in Korea
- Detection and molecular characterization of infectious bronchitis virus isolated from recent outbreaks in broiler flocks in Thailand
- Detection of Antibodies to Infectious Bursal Disease Virus (IBDV) by Agar Gel Immunodiffusion using Recombinant VP2 Protein