Yonsei Med J.
2014 Mar;55(2):449-458.
Influence of Bacterial Presence on Biofilm Formation of Candida albicans
- Affiliations
-
- 1Department of Microbiology, Yonsei University Wonju College of Medicine, Wonju, Korea. leekh@yonsei.ac.kr
- 2Department of Obsterics and Gynecology, Yonsei University Wonju College of Medicine, Wonju, Korea.
Abstract
- PURPOSE
Candida albicans is an opportunistic pathogen that is commonly found in human microflora. Biofilm formation (BF) is known as a major virulence factor of C. albicans. The aim of this study was to examine the influence of bacterial presence on biofilm formation of C. albicans.
MATERIALS AND METHODS
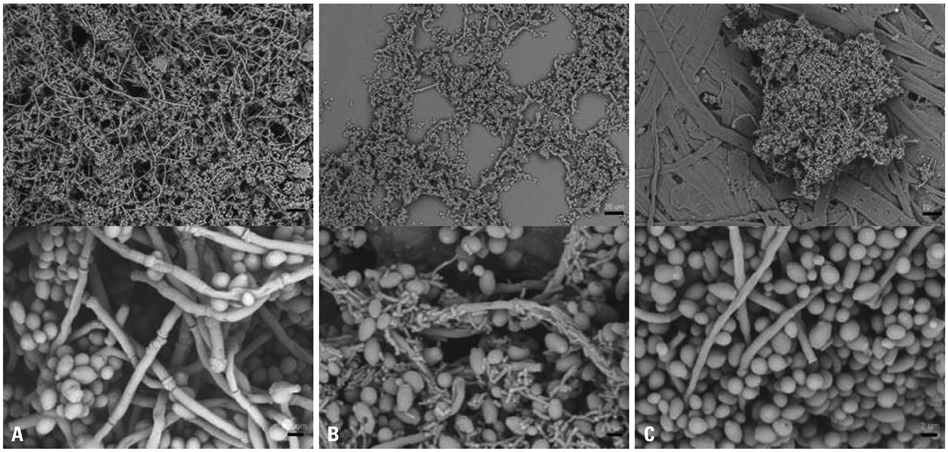
The BF of Candida was investigated when it was co-cultured with C. albicans (C. albicans 53, a yeast with a low BF ability, and C. albicans 163, a yeast with high BF ability) and bacteria. BF was assessed with XTT reduction assay. A scanning electron microscope was used to determine the structure of the biofilm, and real-time reverse transcriptase polymerase chain reaction was used to amplify and quantify hyphae-associated genes.
RESULTS
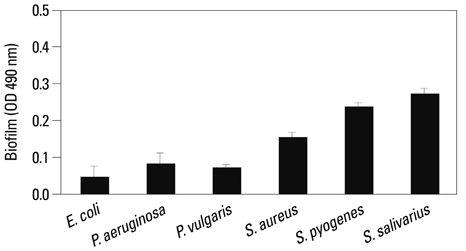
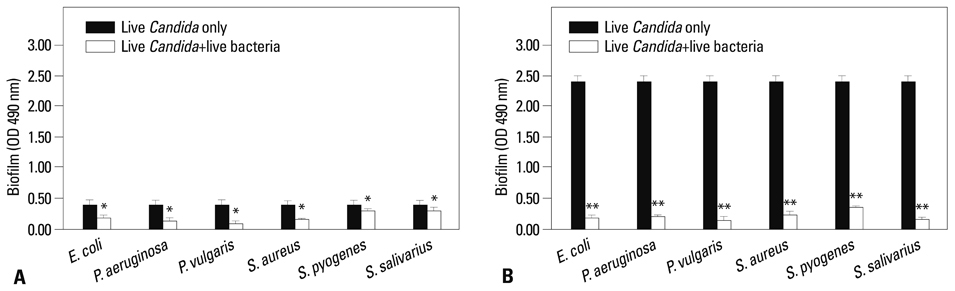
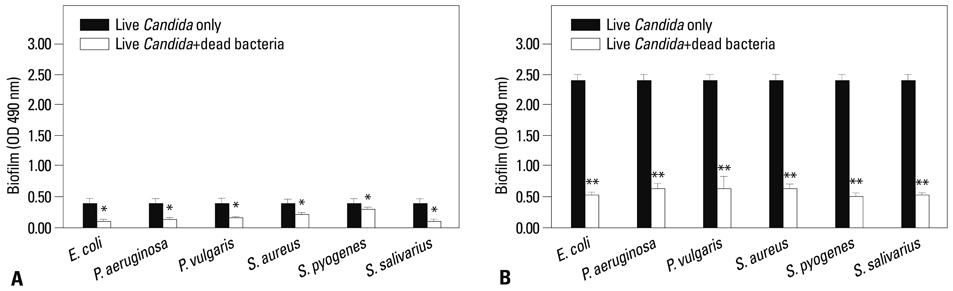
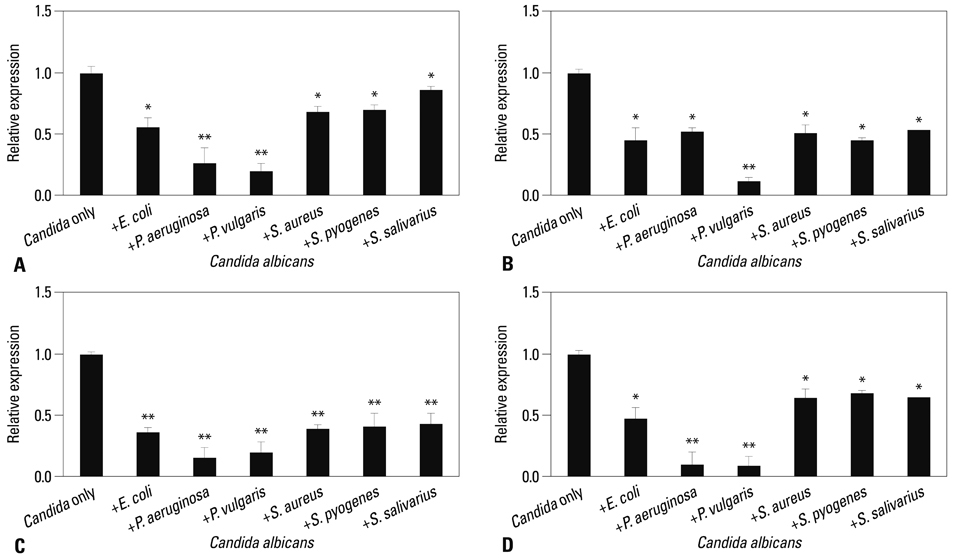
Co-culturing with two different types of bacteria increased the BF value. Co-culturing with C. albicans 53 and 163 also increased the BF value compared to the value that was obtained when the C. albicans was cultured individually. However, co-culturing with bacteria decreased the BF value of C. albicans, and the BF of C. albicans 163 was markedly inhibited. The expression of adherence and morphology transition related genes were significantly inhibited by co-culturing with live bacteria.
CONCLUSION
Bacteria have a negative effect on the formation of biofilm by C. albicans. This mechanism is the result of the suppression of genes associated with the hyphae transition of C. albicans, and bacteria particles physically affected the biofilm architecture and biofilm formation.
Keyword
MeSH Terms
Figure
Reference
-
1. Samaranayake LP. Essential microbiology for dentistry. 3rd ed. New York: Churchill Livingstone Elsevier;2006.2. Lynch AS, Robertson GT. Bacterial and fungal biofilm infections. Annu Rev Med. 2008; 59:415–428.
Article3. Ramage G, Martínez JP, López-Ribot JL. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 2006; 6:979–986.
Article4. Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001; 183:5385–5394.
Article5. Nett J, Andes D. Candida albicans biofilm development, modeling a host-pathogen interaction. Curr Opin Microbiol. 2006; 9:340–345.
Article6. Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999; 48:671–679.
Article7. Kojic EM, Darouiche RO. Candida infections of medical devices. Clin Microbiol Rev. 2004; 17:255–267.8. Hansen SK, Rainey PB, Haagensen JA, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007; 445:533–536.
Article9. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005; 43:5721–5732.
Article10. Manson JM, Rauch M, Gilmore MS. The commensal microbiology of the gastrointestinal tract. Adv Exp Med Biol. 2008; 635:15–28.
Article11. Mclean RJC, Banes MB, McGowin CL, Aron GM. Methods of studying biofilms. In : Ghannoum MA, O'Toole GA, editors. Microbial biofilms. Washington, DC: ASM Press;2004. p. 379–413.12. Tolker-Nielsen T, Hansen SK. Microbial interaction in mixed-species biofilms. In : Ghannoum MA, O'Toole GA, editors. Microbial biofilms. Washington, DC: ASM Press;2004. p. 206–221.13. Shirtliff ME, Peters BM, Jabra-Rizk MA. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett. 2009; 299:1–8.14. Ramage G, Vandewalle K, Wickes BL, López-Ribot JL. Characteristics of biofilm formation by Candida albicans. Rev Iberoam Micol. 2001; 18:163–170.15. Thein ZM, Seneviratne CJ, Samaranayake YH, Samaranayake LP. Community lifestyle of Candida in mixed biofilms: a mini review. Mycoses. 2009; 52:467–475.16. El-Azizi MA, Starks SE, Khardori N. Interactions of Candida albicans with other Candida spp. and bacteria in the biofilms. J Appl Microbiol. 2004; 96:1067–1073.
Article17. Uppuluri P, Dinakaran H, Thomas DP, Chaturvedi AK, Lopez-Ribot JL. Characteristics of Candida albicans biofilms grown in a synthetic urine medium. J Clin Microbiol. 2009; 47:4078–4083.
Article18. Nailis H, Kucharíková S, Ricicová M, Van Dijck P, Deforce D, Nelis H, et al. Real-time PCR expression profiling of genes encoding potential virulence factors in Candida albicans biofilms: identification of model-dependent and -independent gene expression. BMC Microbiol. 2010; 10:114.
Article19. Costerton JW, Montanaro L, Arciola CR. Biofilm in implant infections: its production and regulation. Int J Artif Organs. 2005; 28:1062–1068.
Article20. Li X, Yan Z, Xu J. Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology. 2003; 149(Pt 2):353–362.
Article21. Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science. 2002; 296:2229–2232.
Article22. Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004; 54:1212–1223.
Article23. Bandara HM, Yau JY, Watt RM, Jin LJ, Samaranayake LP. Escherichia coli and its lipopolysaccharide modulate in vitro Candida biofilm formation. J Med Microbiol. 2009; 58(Pt 12):1623–1631.
Article24. Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999; 283:1535–1538.
Article25. Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. 2001; 9:176–180.
Article26. Birse CE, Irwin MY, Fonzi WA, Sypherd PS. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect Immun. 1993; 61:3648–3655.
Article27. Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003; 67:400–428.
Article28. Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003; 67:400–428.
Article29. Armbruster CE, Hodges SA, Mobley HL. Initiation of swarming motility by Proteus mirabilis occurs in response to specific cues present in urine and requires excess L-glutamine. J Bacteriol. 2013; 195:1305–1319.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Influence of Cell Surface Hydrophobicity on Adhesion and Biofilm Formation in Candida albicans and Several Bacterial Species
- Candida albicans Biofilm Formation and Pathophysiology
- Proteus vulgaris and Proteus mirabilis Decrease Candida albicans Biofilm Formation by Suppressing Morphological Transition to Its Hyphal Form
- Biofilm-forming ability and adherence to poly-(methyl-methacrylate) acrylic resin materials of oral Candida albicans strains isolated from HIV positive subjects
- Anti-biofilm Properties of
Peganum harmala againstCandida albicans