Yonsei Med J.
2016 Mar;57(2):298-305. 10.3349/ymj.2016.57.2.298.
The Expression of MicroRNA-155 in Plasma and Tissue Is Matched in Human Laryngeal Squamous Cell Carcinoma
- Affiliations
-
- 1Department of Otolaryngol Head Neck Surgery, General Hospital of Tianjin Medical University, Tianjin, China.
- 2Department of Otolaryngol Head Neck Surgery, Tianjin First Central Hospital, Tianjin, China. dr_shiwenjie@hotmail.com
- KMID: 2374032
- DOI: http://doi.org/10.3349/ymj.2016.57.2.298
Abstract
- PURPOSE
Tumor-associated microRNAs have been detected in cancer, though whether plasma microRNA-155 (miR-155) could be a potential biomarker for laryngeal squamous cell carcinoma (LSCC) prognosis is unclear. We aimed to determine how miR-155 can be used to predict the clinical characteristics of patients with LSCC and correctly diagnose them.
MATERIALS AND METHODS
We collected tissue samples and peripheral blood samples before and after treatment from 280 LSCC cases and 560 controls. Real-time quantitative reverse transcription PCR was employed in this study to compare the relative expression of miR-155.
RESULTS
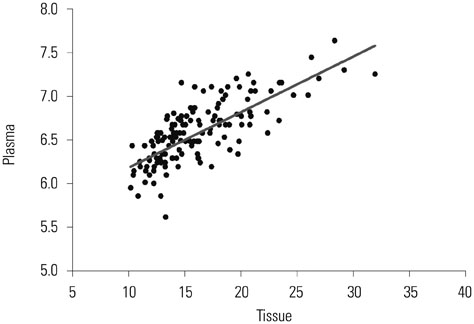
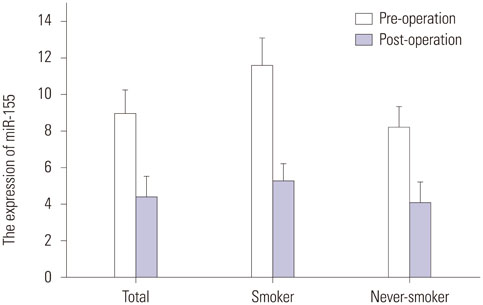
A total of 280 LSCC patients and 560 age- and sex-matched controls were included in the study. The miR-155 level was more up-regulated in LSCC tissue than in the non-tumor tissues (13.6+/-2.4 vs. 3.1+/-0.80, p<0.001). Additionally, a significantly higher miR-155 level in plasma samples from LSCC patients than in those of the controls (8.9+/-1.25 vs. 1.8+/-0.8, p<0.001) was reported. Tissue miR-155 showed an area under the curve (AUC) of 0.933, with a sensitivity of 82.6% and a specificity of 89.2%. The AUC for plasma miR-155 was 0.757, with a sensitivity of 58.4% and a specificity of 69.5%. When early LSCC in TNM I stage was considered, tissue miR-155 showed an area under the curve of 0.804, with a sensitivity of 85.2% and a specificity of 87.3%.
CONCLUSION
The expression of tissue and plasma miR-155 were significantly up-regulated in patients with LSCC. Our work will serve as a basis for further investigation, preferably large-scale validation in clinical trials.
MeSH Terms
-
Aged
Biomarkers, Tumor/blood/*genetics
Carcinoma, Squamous Cell/blood/diagnosis/*genetics
Case-Control Studies
Early Diagnosis
Female
Gene Expression Profiling
Gene Expression Regulation, Neoplastic
Head and Neck Neoplasms
Humans
Laryngeal Neoplasms/blood/diagnosis/*genetics
Male
MicroRNAs/*blood
Middle Aged
Prognosis
Real-Time Polymerase Chain Reaction
Up-Regulation
Biomarkers, Tumor
MicroRNAs
Figure
Reference
-
1. Gourin CG, Starmer HM, Herbert RJ, Frick KD, Forastiere AA, Eisele DW, et al. Short- and long-term outcomes of laryngeal cancer care in the elderly. Laryngoscope. 2015; 125:924–933.
Article2. Landry D, Glastonbury CM. Squamous cell carcinoma of the upper aerodigestive tract: a review. Radiol Clin North Am. 2015; 53:81–97.3. Rutt AL, Hawkshaw MJ, Sataloff RT. Laryngeal cancer in patients younger than 30 years: a review of 99 cases. Ear Nose Throat J. 2010; 89:189–192.
Article4. Park JM, Jung CK, Choi YJ, Lee KY, Kang JH, Kim MS, et al. The use of an immunohistochemical diagnostic panel to determine the primary site of cervical lymph node metastases of occult squamous cell carcinoma. Hum Pathol. 2010; 41:431–437.
Article5. Qiu G, Li Y, Liu Z, Wang M, Ge J, Bai X. Clinical value of serum HMGB1 in diagnosis and prognosis of laryngeal squamous cell carcinoma. Med Oncol. 2014; 31:316.
Article6. Vasca V, Vasca E, Freiman P, Marian D, Luce A, Mesolella M, et al. Keratin 5 expression in squamocellular carcinoma of the head and neck. Oncol Lett. 2014; 8:2501–2504.
Article7. Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015; 518:107–110.
Article8. Loh YH, Yi SV, Streelman JT. Evolution of microRNAs and the diversification of species. Genome Biol Evol. 2011; 3:55–65.
Article9. Ceppi P, Peter ME. MicroRNAs regulate both epithelial-to-mesenchymal transition and cancer stem cells. Oncogene. 2014; 33:269–278.
Article10. Xu TP, Zhu CH, Zhang J, Xia R, Wu FL, Han L, et al. MicroRNA-155 expression has prognostic value in patients with non-small cell lung cancer and digestive system carcinomas. Asian Pac J Cancer Prev. 2013; 14:7085–7090.
Article11. Lind EF, Ohashi PS. Mir-155, a central modulator of T-cell responses. Eur J Immunol. 2014; 44:11–15.
Article12. Zheng Y, Xiong S, Jiang P, Liu R, Liu X, Qian J, et al. Glucocorticoids inhibit lipopolysaccharide-mediated inflammatory response by downregulating microRNA-155: a novel anti-inflammation mechanism. Free Radic Biol Med. 2012; 52:1307–1317.
Article13. Bera A, VenkataSubbaRao K, Manoharan MS, Hill P, Freeman JW. A miRNA signature of chemoresistant mesenchymal phenotype identifies novel molecular targets associated with advanced pancreatic cancer. PLoS One. 2014; 9:e106343.
Article14. Chen L, Jiang K, Jiang H, Wei P. miR-155 mediates drug resistance in osteosarcoma cells via inducing autophagy. Exp Ther Med. 2014; 8:527–532.
Article15. Pareek S, Roy S, Kumari B, Jain P, Banerjee A, Vrati S. MiR-155 induction in microglial cells suppresses Japanese encephalitis virus replication and negatively modulates innate immune responses. J Neuroinflammation. 2014; 11:97.
Article16. Zhu J, Chen T, Yang L, Li Z, Wong MM, Zheng X, et al. Regulation of microRNA-155 in atherosclerotic inflammatory responses by targeting MAP3K10. PLoS One. 2012; 7:e46551.
Article17. Anzi S, Finkin S, Shaulian E. Transcriptional repression of c-Jun's E3 ubiquitin ligases contributes to c-Jun induction by UV. Cell Signal. 2008; 20:862–871.
Article18. Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014; 15:429.19. Ayaz L, Görür A, Yaroğlu HY, Ozcan C, Tamer L. Differential expression of microRNAs in plasma of patients with laryngeal squamous cell carcinoma: potential early-detection markers for laryngeal squamous cell carcinoma. J Cancer Res Clin Oncol. 2013; 139:1499–1506.
Article20. Sun X, Song Y, Tai X, Liu B, Ji W. MicroRNA expression and its detection in human supraglottic laryngeal squamous cell carcinoma. Biomed Rep. 2013; 1:743–746.
Article21. Wu TY, Zhang TH, Qu LM, Feng JP, Tian LL, Zhang BH, et al. MiR-19a is correlated with prognosis and apoptosis of laryngeal squamous cell carcinoma by regulating TIMP-2 expression. Int J Clin Exp Pathol. 2013; 7:56–63.22. Jones K, Nourse JP, Keane C, Bhatnagar A, Gandhi MK. Plasma microRNA are disease response biomarkers in classical Hodgkin lymphoma. Clin Cancer Res. 2014; 20:253–264.
Article23. Vimalraj S, Miranda PJ, Ramyakrishna B, Selvamurugan N. Regulation of breast cancer and bone metastasis by microRNAs. Dis Markers. 2013; 35:369–387.
Article24. Gao F, Chang J, Wang H, Zhang G. Potential diagnostic value of miR-155 in serum from lung adenocarcinoma patients. Oncol Rep. 2014; 31:351–357.
Article25. Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010; 16:1129–1139.
Article26. Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, et al. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014; 5:1174–1184.
Article27. Kopp KL, Ralfkiaer U, Nielsen BS, Gniadecki R, Woetmann A, Ødum N, et al. Expression of miR-155 and miR-126 in situ in cutaneous T-cell lymphoma. APMIS. 2013; 121:1020–1024.
Article28. Sochor M, Basova P, Pesta M, Dusilkova N, Bartos J, Burda P, et al. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer. 2014; 14:448.
Article29. Sun J, Shi H, Lai N, Liao K, Zhang S, Lu X. Overexpression of microRNA-155 predicts poor prognosis in glioma patients. Med Oncol. 2014; 31:911.
Article30. Zhao XD, Zhang W, Liang HJ, Ji WY. Overexpression of miR-155 promotes proliferation and invasion of human laryngeal squamous cell carcinoma via targeting SOCS1 and STAT3. PLoS One. 2013; 8:e56395.31. Xu L, Li M, Wang M, Yan D, Feng G, An G. The expression of microRNA-375 in plasma and tissue is matched in human colorectal cancer. BMC Cancer. 2014; 14:714.
Article32. Zhang R, Wang W, Li F, Zhang H, Liu J. MicroRNA-106b~25 expressions in tumor tissues and plasma of patients with gastric cancers. Med Oncol. 2014; 31:243.
Article33. Herberth G, Bauer M, Gasch M, Hinz D, Röder S, Olek S, et al. Maternal and cord blood miR-223 expression associates with prenatal tobacco smoke exposure and low regulatory T-cell numbers. J Allergy Clin Immunol. 2014; 133:543–550.
Article34. Basso K, Schneider C, Shen Q, Holmes AB, Setty M, Leslie C, et al. BCL6 positively regulates AID and germinal center gene expression via repression of miR-155. J Exp Med. 2012; 209:2455–2465.
Article35. Sandhu SK, Volinia S, Costinean S, Galasso M, Neinast R, Santhanam R, et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Eµ-miR-155 transgenic mouse model. Proc Natl Acad Sci U S A. 2012; 109:20047–20052.
Article36. Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005; 102:3627–3632.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of Cyclooxygenase 1 and 2 in Laryngeal Squamous Cell Carcinoma
- Expression of bFGF and CD-31 in Laryngeal Squamous Cell Carcinoma
- An Immunohistochemical Study on the Expression of Matrix Metalloproteinase-2, Tissue Inhibitor of Metalloproteinase-2 and Angiogenesis in Human Laryngeal Squamous Cell Carcinoma
- Expression of p53 and MHC Class I According to HPV Infection in Laryngeal Squamous Cell Carcinomas
- Detection of Epstein-Barr Virus in Laryngeal Squamous Cell Carcinoma